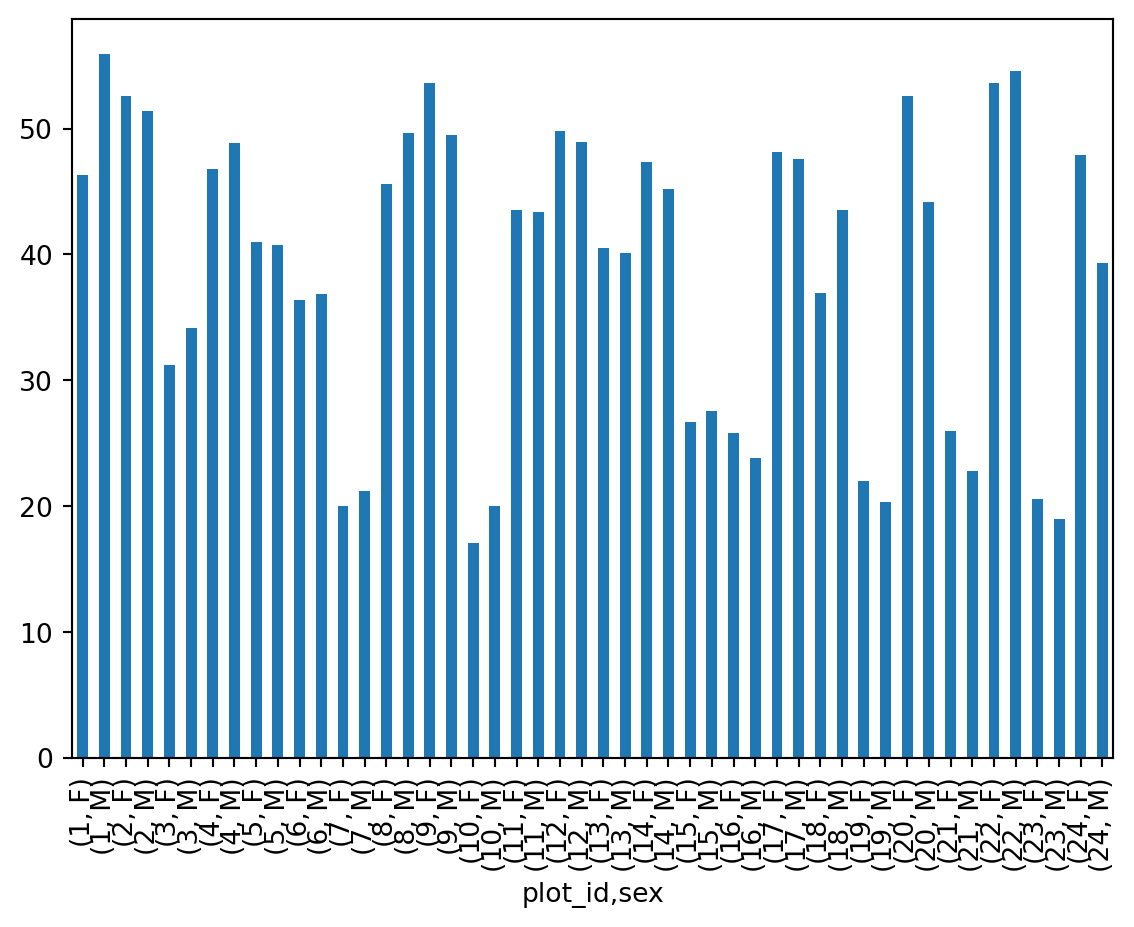
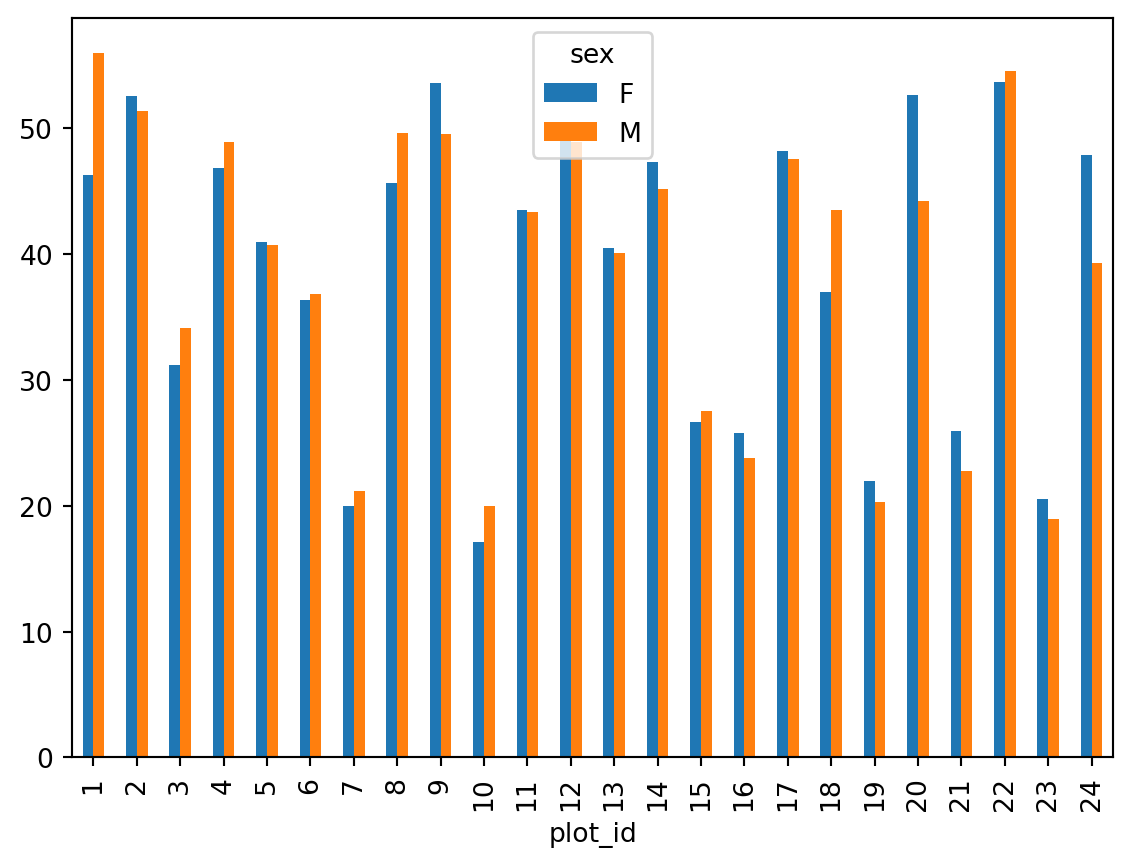
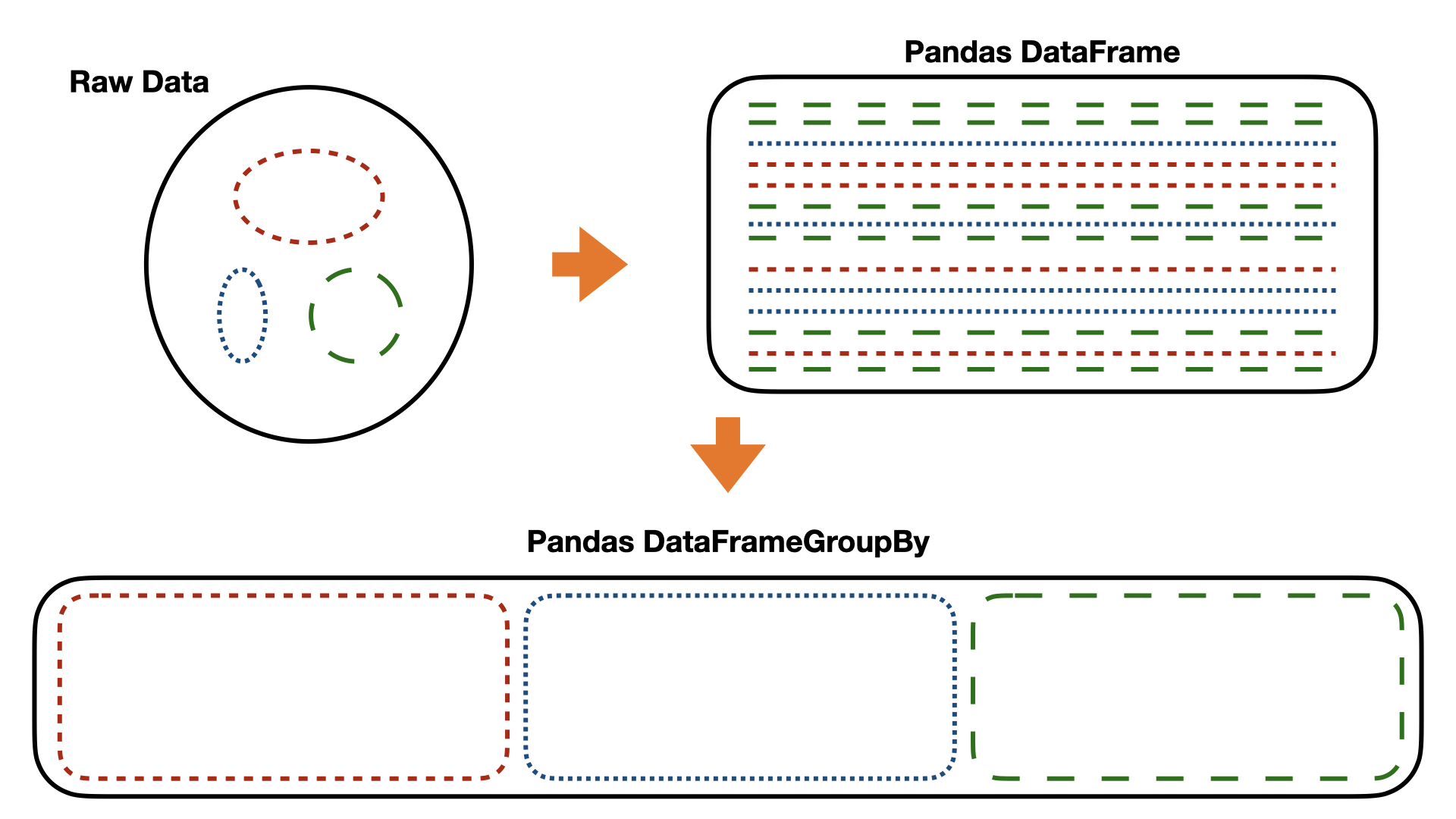
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/groupby.py:1942, in GroupBy._agg_py_fallback(self, how, values, ndim, alt)
1941 try:
-> 1942 res_values = self._grouper.agg_series(ser, alt, preserve_dtype=True)
1943 except Exception as err:
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/ops.py:864, in BaseGrouper.agg_series(self, obj, func, preserve_dtype)
862 preserve_dtype = True
--> 864 result = self._aggregate_series_pure_python(obj, func)
866 npvalues = lib.maybe_convert_objects(result, try_float=False)
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/ops.py:885, in BaseGrouper._aggregate_series_pure_python(self, obj, func)
884 for i, group in enumerate(splitter):
--> 885 res = func(group)
886 res = extract_result(res)
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/groupby.py:2454, in GroupBy.mean.<locals>.<lambda>(x)
2451 else:
2452 result = self._cython_agg_general(
2453 "mean",
-> 2454 alt=lambda x: Series(x, copy=False).mean(numeric_only=numeric_only),
2455 numeric_only=numeric_only,
2456 )
2457 return result.__finalize__(self.obj, method="groupby")
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/series.py:6549, in Series.mean(self, axis, skipna, numeric_only, **kwargs)
6541 @doc(make_doc("mean", ndim=1))
6542 def mean(
6543 self,
(...) 6547 **kwargs,
6548 ):
-> 6549 return NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/generic.py:12420, in NDFrame.mean(self, axis, skipna, numeric_only, **kwargs)
12413 def mean(
12414 self,
12415 axis: Axis | None = 0,
(...) 12418 **kwargs,
12419 ) -> Series | float:
> 12420 return self._stat_function(
12421 "mean", nanops.nanmean, axis, skipna, numeric_only, **kwargs
12422 )
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/generic.py:12377, in NDFrame._stat_function(self, name, func, axis, skipna, numeric_only, **kwargs)
12375 validate_bool_kwarg(skipna, "skipna", none_allowed=False)
> 12377 return self._reduce(
12378 func, name=name, axis=axis, skipna=skipna, numeric_only=numeric_only
12379 )
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/series.py:6457, in Series._reduce(self, op, name, axis, skipna, numeric_only, filter_type, **kwds)
6453 raise TypeError(
6454 f"Series.{name} does not allow {kwd_name}={numeric_only} "
6455 "with non-numeric dtypes."
6456 )
-> 6457 return op(delegate, skipna=skipna, **kwds)
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/nanops.py:147, in bottleneck_switch.__call__.<locals>.f(values, axis, skipna, **kwds)
146 else:
--> 147 result = alt(values, axis=axis, skipna=skipna, **kwds)
149 return result
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/nanops.py:404, in _datetimelike_compat.<locals>.new_func(values, axis, skipna, mask, **kwargs)
402 mask = isna(values)
--> 404 result = func(values, axis=axis, skipna=skipna, mask=mask, **kwargs)
406 if datetimelike:
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/nanops.py:720, in nanmean(values, axis, skipna, mask)
719 the_sum = values.sum(axis, dtype=dtype_sum)
--> 720 the_sum = _ensure_numeric(the_sum)
722 if axis is not None and getattr(the_sum, "ndim", False):
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/nanops.py:1701, in _ensure_numeric(x)
1699 if isinstance(x, str):
1700 # GH#44008, GH#36703 avoid casting e.g. strings to numeric
-> 1701 raise TypeError(f"Could not convert string '{x}' to numeric")
1702 try:
TypeError: Could not convert string 'DMPEDMPFDSDMDMDSDSDMNLDSDMDMDMDMOTDMDMDMDSDMDMPPDMDMDMDMDMDOOXDMDSDMDSDMPFDMDMDMDMDMPFDMPPDSDMDSDMDMDMDMPFDMDMOXDMDMDMDMDMDSDMPFDMDMDMDMDSDSDMPPDMDSDMPFDMOXDMOXDMDSDMDMPFDSDMDMDSDSDMDMDSDMDMPFPPDSDMDSDSDSDMDMDSDSDMDMDMDSDSDMDMDSOLDSPEDMDSDODMDODSDSDMDMDSDMDODMOLDSDODSDMDSDSDMDODSDMDMDMDMDODSPFDMDMDSDMPFDMDMPFPFDSDMDSDSDMDSDSDSDMRMDSDMDSPFDMDSDODMDMDSDMDMDMDOPFPFDMDMDMPFDSDMDSDSDODSPFDSDSDMDSDMDMDMDSDMDSDMDMPFDSDODMDSDMPFDMDSDMDMDSDSDSDSDMDMDSDSDMDMDMDSDMDMDMPFDMDSDMOTOTDMDMDMDSOTDSDSDSDMDSDSDMOLOTDMDMDMDMDSDMDSDMPFDSDMDSOTOTPFDMDMDSOTPEDSDSPFDSDSDSDSDMDMDMDMDSOTDMDMDMDMDSDMOLOTDSDMDSOTDMPEDMDMPFDSDMNLOTDMDSDSDMDMDMDSDSDSDMDSDODSDMDMDSDSDSDSDMDSDSDMDMDSDMDSDSDMDMDSDMDSDSDMDSDSDSDSDSDSDMOTOLDSDMDMDSNLDSDSDSNLDSDMPMNLDMDSDSDSDMDSDMDMDSDSDMDMDSDMOTDSOTPFDSDSDSDSDMPPDSDSDSDSDMDMDSDOOLDMDSDSDMDMDSDSDSDSDMNLDSDSDSDMPFDSDMDSDSDMDSDMDSDMDMOTDSDSDSDSDMDSDSOTDMDSDMDSDMDMDSDSDSDSDMDSDSDMDSDMDSDMDSDMDMDSOTOLDSPPPPDMDMNLDMDSDMDSDMDMDMDSDSDSDSDMDSDSPEDSDMDSDSDMDMPPDSNLPMDMDMDSDSDMNLDMDMDMDMDSDMDSDMDSPFDSDSDMNLDSDMOLDMDSPPDSDMDMDSDSOLDSDSOLNLPEDMDSDMDMDSDSDSDSNLPPDSDSDMDMDSOTDSDSPEDMDSDSNLDMDMDMPPDSDMDSPPPFDSDMDMDMDSDSPFDMDSPFPEOTDSDMDMDMDSDSPEDSDSDSDMDMDMDSDMDSDMDMDMDSPPDMDMDSDMDSDMDMDSDSDSDSDSDSDSPFDMDMPPDMOTDMPFDMDSPFDSPPDMPFDSDSPFOTDMDMDSOLPFDSDMDMDSDSDSDMDSOTDMDMDSOLDMDMDMDODMOTDSDODSPFDSOLOTDMDMDMDMDMDSDSDSDSDMOTDSDMDMDMDMDMOTOLDMOLDMDSDMDMDSDMOTRMOLDMDSDSDSDSDSOLDODSNLDSDSDMDMDMDSDSOLOTDMDMDMDSOLDMOTDMDSDMDMDMDMOLDMDMDMDSDMOTDMDSDMDMDMOLOLDSDMDMDMDMDMDMOLDMOLDODSDSDSDODSDSDSDMOLDMOTDSPMDSPMDSDMOLDMDMOLDMDSDMDSDSDMDSDSPFDOPFDMDSDSDSDODMDSDSDSDODSDSDSDSNLDMNLDSDMDSDSDSDSDSDSDSDSDOPPDONLDODMDMDSDSDSDMDSDMDSDSDSPFDSDSDSDMDSDSDSDMDSOTPPDSDMDODSDSDMDSDSDMDMDMOTPFDSDMPFDSNLDSDSPFPEDMDMOLPPDMDMDSDSDSDSDSDMPPDOPEOTDMDMDSNLPFDSDSPFOTDMDSDSDSDODSOLOTPPDMPPDMOLPPPPDSDSDMOLDSOTNLDSOLDMDMDSDSNLOTPPDMNLDOOTDSOTDSDMDSOTPPOTOLDSNLDMOTDMDSDSDSPEPPDMOLOLPPDMOTPFOLDMOTDSDMDMPFDSDMDMOTOLOTPPNLDSNLDODMDMDSOLDMOTDSDSOTDSOLDSDSOTOTDMDMDSPPDMOTDMNLOTPEDMDMOTOTOTRMOTOTDSOLDMPFDSOLDMOTNLDSOLPFDMNLDMDMDMDMDODSDMOTDMDONLDSNLDMDMDMOLDSOLDMNLDMDMOTDMNLDSDMOLDSRMDMRMDMPEDSPFDMDODMOTDMOLRMDSOLDMDSDMDODMDSRMOLNLOTOLNLOLRMDOPEDSPFDMDMDMDMDMDMDSDMOTDMOTDMDMDMDMDMDMDOOLDMDMRMDSOLDODMDSDMDMOLOTPFDMNLDSDMDMDMPFPFNLOTDMDMDODMOTPFOLRMDORMOTOLDORMDMDMPEDSOLDMDODODMDMDMDMDMDODMDMDMOTDMDMDMDMDMDMDMDMDMDMDMNLDSOLPFOTPFDMRMNLRMDSDMOTPEOLDMPFDODOOTOTDSDMPFDMDMDMDSDMDMDOOLDMOLDMPERMDODMPFDMRMDMDMDMDMOTDMDMDSOTDMDMDMDOPFOTDSDSRMDODODMOLDMOLDMDSDMDSDMOLDMOTRMDMOTRMDMDORMDMDMDMDOOTPEDSDMDMDODMDODMDMNLPFDMDMDMDMDMDMDMDMDMOLOLPFRMRMDMRMRMDMDSDSRMDSDMDSPEDSDMOTDMDMOTDSDSDSDSPFOTDMDSPFDMOLDSPFOTDMDSDSDODMRMOLPPDSDSOLDMPFOLOTDMOTDSDMRMDSRMDMDSDMOLNLDSDMDMDSDMDSPEDSPFDMDSDMDSDSDSOTDSDODMPPDSOLDSOLDSDMDSDMDSDMPFPFNLDODSNLNLNLDMDSDSDSDSDMDSDSDSDSDMDMOTDSDSNLDMDMOLDMDSDSDSDODMDSNLDOPPDODSDSDSDSDSDMOLDMNLDMPPDMDMDSDMDMDMDSOTOTDMNLPEOTNLDSDSDSDMDODODMNLDMDMDMPPPFDSPFDMOLDMNLOLDSDSNLNLPPDMPFDMDMPFPPDMOTDMPFDSDMDSNLPPOTPPOLDSDSPFDSDMDSPEDSPFPPDMNLPPDMDMPFDSOTPFDMDMPPDSPFOTOLPPDSRMOTDSDSDMOTOTDSPFPFOTDOPFDMOTOLDMNLDSDSDSDSDMOTOLDMOTPPDMPENLOTDODMOTDSDSNLDODSDSDSPFPFDOOLDMDSOLDMDMDODMOTOTDMDSDSDMDSDMDSDSDMDMDMDSOTDMDMDMDMDMDODMDMOTDSOLOLNLDMDMOLDSDMDSPFDSOTDMDSOLPFOLDMOTDSDMDMDMDMNLOTOLOLOLDSOTDMDSDMDSPFDODMOTDMPFDSDODODSDMDSDMDMDMDSDSDMDMDMDMDMDMDMPFNLNLDMDODMDMDSDSDSDSNLDSDSDOOTDMPFOLDMDMPEPEDMNLNLDODOOLPPRMDSDODSDMOTDSDSDMDODMOLDSOTOLDMDSDMDMDMNLDMDMDODMDMPMDSDMDMDSDMDSOLDMPFNLDMOLDSDMDODSOLDMDMOLDOPENLOLNLPEDSDSOLDMPFPPDODSDSDSRMDSDMOLDSDODMDMDSDMDMPFDSDMDMDODMDODSDODOOLDODMDODSOLRMOTDSDODMDMDODSOTDSDSDMDSDSDODMOTOLDODMDMDMDMDSDMDMDSDSDSOTDMDMDSPFDMDMPFOLDMDSDMDSPFPFDSDMDMDMDSDMDSDMDMDOOLDODSDMDSDMOLDSDMDMDMPEDODMDODMOLPFNLDMPPDMDODMDSDSDSDODSDMDMDMPMDMDODMDMDSOLPFDMRMOLDSDMDMDMDMDMDMDSDMDMDMDMDSDSDSDODSDMDMDMDMPFDMOTPFDSDODODSDMDSDSDMDODSDMDODMDODSDMDMOLDMDMDSDMDMDSDODMDMDSDSDOOLDMDMDMDMDSDMDSDMDMDODODSPPDMDODMDMDSDSDMOLPPDMOTDODMDODMDMDMDMDODSDMDODMDSPEDSDMPPDMNLPMDMDSRMDMDMDMDSDSDSDMNLDMDSDMDODMOTDMNLDODMDMOTDMDODSDMDODSDMDSDMDMDSDMDMDMDODSDSDODSPFDMDODODSDMDSDSDSPFDMDMDMDMDODODSDSDSDSDMPPDSDODSDSPPDSRMOLDSNLDMPMDSDMDSDMDMDMPEDMNLDMPPPFDMDMDMDSDSDSOTDSDSDMOTDSDODMDMDONLDMDODMDMDMDMDMPPDSDODMDMDODMDODSDOPFPFPFDMDSDSNLDSDSDMDSDSNLDMDODMDMDSDMDMOTDSDSDMDSOLDODSDMPPPPDMOTDMOTDODMDMDMPPPPDMDMOLPFDODMNLDMDSDODSDMDSDSDODMDONLDMDMDMDMDMDODMDSOTDSDMOLNLOLDMOTNLDSDMDMNLDMOTDSDSDSDODSSSDMDMOLDMDMDMDSDMPEDMOLOTOLDMNLOTDMDSOTDMDMDMNLPFPFDMDSOLDMDMDOOTPFDODSDODSDMDSOTOTDMNLDMDMPEDMDMDMDSDSDONLOLDMOLDSDMOTDSOTPMRMOTPFNLDSDSRMDONLDMDMDODSDMDMDMDMPFDMDMOLDORMDMDMPEPERMPMDSPEPFRMPEDMRMRMPEDSDMDMRMNLPMDSDMPFRMPENLDMDSDMPEPFPEDMRMDSDMDODSDMDMDODMRMNLRMNLDSDMDSDOPEDORMDMPEOLRMDORMDMNLDODMRMPEDSDSRMDMDMPEDMDODSPMNLRMDSDMDMDMDODMOTOTDMRMNLPEDSPEDMNLDMDMDMPFRMDSOLPFNLPFRMDMDMDMDMPERMPFDMDMDSNLDMRMPEPFDMPEPMDSRMPFPEDMNLPFRMRMDMRMDMPENLDMDSRMDMNLPEDMDSRMDMRMDMDMPFPFOLDMRMDMRMRMDMOTOLDMDSDSPEDSDORMDSPEPEDSDODSOTOLOLDODSDSNLDSOTRMRMRMRMRMDMPEPEDSPFPEPFRMPEDMRMRMPFDMDODMPMRMDODMRMDMDSPMDMDMPEDMDSNLDODMDONLPFPFDMDSDSOTDMDMNLDSOLPMOLRMPFRMPMDMRMDMDMDSDSDMPEDODMPMDMPPDONLDSPPDSPFDMDMPERMPMPEDSDOPEDMDODMDSPENLPMRMDMPEDODMRMDMDSPPDSDMDSNLNLPMDSDSDSDODODSDSDMRMDSDMDMPFDMPEDSDMDSPFOTDSDSNLDMRMDSDSDMDODMDSDMDODSDSDMDMNLPPPEDMDSDMPMDMDMNLDMPPOTOLDMPFDMNLPFNLDSDSDSDMDSDODMDODSDSPEPFRMNLPPDSOTDMDMDMOLDSDMDMOLPEPFDMRMDODSPMDSDMPFDMRMNLNLDSDMNLPEDSNLDMDSOTNLDMNLDSOTPPOTDSDOPEDSSHDMDMPMDMPERMDMDMDODONLDSDMOTNLDSDSDMDMNLPFPPDMDMDMPMPPDODMDOPPDSOLOLDMPPPEDOOLDMOTOTDSNLOTOTPFDSDMPFPENLDMDSDSDMDMPFDSDSDMDSDMNLOTPPPMDMDMNLDSPPPFDMDODMPPPMDMPFDMDMDMPFDMNLDODMDMPMNLNLNLPMDMDMNLPPDONLRMDSDSPMPFDSDMNLPENLNLOTDMPMPMDSDMDOPPDMPPPFPFOTDMDSDMDMPPDMDMDMDMDSOTPFNLDSOTDOOTDMNLDMDMDSPEDOPMDMRMNLDMDMDOPFPEPMDSPPDMDSPFPEDMPFDMPPDODMDMDMDSNLRMDMNLDMDSDMNLDMRMDSPPOLNLOTDMPFOLDMDMDSOTDSDMDMOTDMNLOTDMDMDONLDSDMDSOLDMDSDSDSPPDMDMDSDMDSDMDMOTDMDMOTPPPFDSDODOPEDMPFOTPFNLPPDMDONLOTDMOTDSPMPPOLOLNLDMDSOTOLDSDONLDODMDSDMDMPFDMRMDMDMDODMDMDMDSDMDSOLDODSPEPFDMDMDMDMDSDMPFOTDMDODSDODSPFOLDMDSDMDMDSDMDMDMDMDSDMOTDMOLPMDSPMOLPEDMDOOLDMDMDSDMDMDMDMOLDMDSDMDSDODSPFDMDSDSPFDSDMDMOTDSNLDMDMDMDMOTOTDMDSDMOTDODSDMDODMDSDSDMDMDSDMDMOTPEDMDMDSOTDMDMDMDSDODODMDMPFRMDOPEDSOTDMOLDMDMDMDMDMPFDSDMDMDMDSDMRMRMNLDMDMNLDOPMOLNLRMDMPFDODSDMDOPFDSNLDSNLDMNLDMPFDMDODMDSDODMOLRMDMDMPFPFDMRMDSDMDMDOOLDMDSPFDODSDSDODMDODSDMDSDMDSOTDMDMPMDMNLDMDMPFDSPMRMRMDOOLOLRMDMDMDMDMDMRMDMRMRMNLPFDORMDMDMDMDMRMRMDMDSDMDMDMRMDSOTDODMDODMOLDODODMDMDODSDMDMDMPFDODMDMDODODMDSDMDODSDMDMDMDMPFDMDMDSDSDMDOOTOLDMRMDMDMOTDOOTRMDMOLDODMDMDMDORMDMDMRMDOOLDODMDORMDMDMRMRMDMDMDSDMDODODSDMDMOLDMDMDMDSDMPEDMDMDMDMDMRMPFDSDMDMDMRMRMDSDMRMRMOLDMPFDMPEDSDMDMOTRMDMDMRMDSDMDMRMDSDMDSDOOTDMOLDOOLDMDMDODOPFRMDSDMDSDODSDOPFDMPFDMDMPFDODSOLDMDMDMDSDMDMOTRMDSRMNLDMDSDSDMRMRMDMDMDSDMDMDMDMRMOLNLRMDMPFPFDMDSPEPFDSDSRMDMPFDSDMDSDSDSPEDMPFDSDODSDMPERMPPDSDSDSRMPFDSDMDSPFDSDSDMDODMDMPPPFRMDSDSDMDMRMOTDOPEDSDSPMDMOTDMRMDSDMRMNLDMNLNLDMRMDMDMPERMDMOTDMDOPEPFPFDMDMDSDMPPNLDSNLDSDSDSPFPPDSDMRMDMRMOTDSDMDONLPPDSDOPMDODODONLDMNLPEDONLOTDMOTDMDODODSDMDMDMRMPFDMOLNLDMDSDSDOOTDOOTDSPPNLPFPFDSDMDSOLNLDSDSDMDSDMDSDSDMOLDMPPOTDMNLNLDMDODMRMDMDMPFNLDOOTDMPMDMDOPFDODODMDMPPDSDSNLDMDMOLDMDMDMDMOTPFNLDSPPDMOLPPDMDSNLOLDMOTDSDMDSNLPPNLDMNLDMDMNLOTDMDMDODMDMDODSPMDMDMDODMPPDMDMDMPPOTOTNLNLDMDONLPPDMPFPFRMDMPFOTNLOTDMDMOTDMDSDSPPPFPPPPDMDMNLDMNLDSDMNLDSDMNLNLDSDOPPDMNLNLNLDMNLDODMDODMDODSDMDMRMPEDMNLDMDMDMDMNLNLDODMOTDODMDMNLPFOLDMPPPPPFDSNLDSDMDMDONLDMRMDSDSOTDODMNLRMDMOLDSNLDSDSDMNLDMDMPPDODMPPPPDOOLDSDMDSDMDSOLNLRMDMDODSDMDSDMDMOTOTOTDMDMDMDMOTDMDMPFDOPENLDMDSDMDSDSDMOTOTDONLDMNLDODMDMDMDSDMDSNLDSNLDSDMDMDMDSDMDSDODMDSDMDMDSNLPPDMDMDMDMNLDMOTDSNLDMNLDMOTDSDMDMDMRMOTPMDODOOTDSDMOLDSDODMOTDMDMDMOTDODMRMDMDOOTDMDMDSNLPFDSDSOTDODSDSPPDSPMDODMDMOLDMNLDMDMDSDMDMNLOTDMDMNLOLDMDSDMDMOTDSRMOTDMPENLDOOLDSRMDMDMOLDMDMPMDMDSDSDMDMDMDSDMNLPFDMNLNLDMDMDODMDSNLDSDSDODSOLDMDODODMDMDMOTDMNLDMNLDMDSDSOTDMDSPEDMDMDMRMOTDMRMDSDODMDMDMDMRMRMOTDMDMDSPMDSOLDMDSDMDMDMDSDMRMDODONLOLDMRMDSDODMDSRMDMDMDSDMDMDODMDMDMDMDSDMRMDMDMOLDSDMDMDSDMOTDMDMDMDSRMDSDMDODSPMDMDMDMDMOLDSDSDMDMDODMDOPPPPDMDODMDSDSNLOTPMDMRMNLDMDSDODSDMDSNLDSDMDMPFDMPPDMOTDSDODOPPPPOLNLDSDMDMOLDMDMDMDMOTDMNLDSDODMDMDMDODMDMPEOTDMDMOLDSDMDODMDMDMDMDSOLDMDMDMDMDODMDMDSOLDMRMDMDODODSOLDODOPMDORMDMDMDMDMDMDODSDMDODMDMDMDMOTDMDMDODMDOOLDMNLOTNLDODSDMDMDORMOLDMNLDSDMDMNLDMDMRMDMDMOLOLPPDMNLDODMDONLDMRMDODMOLDMDMDMDMPERMDMDSDMPENLDMRMDMOLDMDODMDMDORMDOOLNLNLRMPPDMDMPPOLNLDMDODMPERMRMOTNLDMPMOLOLDMNLDMDMDODMOTNLNLDMNLPEPPPPNLDMOLDMOLRMOLDOPEDMNLPPNLDSDMDMDMOTNLOTNLRMOTDMPEDMOLDMDMRMDMDMDMNLDODMDODMOLNLDODMNLDMDMOLDMDMDMRMDSDMDMDMDMRMOTOTDMDSDMDMPHDMDODMDMDMRMDMDMDMOLDMDMDMNLDMDMDMOLDMDODMRMDMDMDMPHDMOLDMDMDMDMOTRMDSOTPHDOOTOLDMDMDSDODOOLDMDMDODMDMDMOLDODODMPHDMDMOLDMDMOLDMOLDMDMOLDMDMDMDMPHDMDODMDORMRMOLDMDMDODMDMDMOTDMOTDMDODMPMDODOPEOTDMDMDMRMOTDODMDMDMDMDMDODOOLDODMDMDMPMDMOLDMOTDOOTDODODMRMDMPMDMDMDMDMDMDMDMDOPEDODODSDMDMOLDMDMDODMRMDORMDMDMDMDMDODMDMOLDMOLDMDODMDMDMDMDMDODOOLDOPERMDOPEDMDMDSDMDMOLNLDMDMDMDMDMDMDMDMDMDOOLDODMPERMDMOLOTDORMOLDMDMDMDMDMDMDMOLOLDMOLOLPMDMDODMDMDODMOLDOPMDMDMNLPEDSDMDOOLDMDODODODSOLDMDMDMDMRMDMDMDMDMDODMDMDMOLRMOLDODMOLDODMDMDMDMDMDMDSDMDMDMDMDMOLPPDMDMDMDOOLDMDMDMDMDMDMDODMDOOLPMDMDMDMDORMDMDSDMDMDODMOLOLDMDODODODMRMDMDSPPDMDMDODOPMOLDODMDMDMPPOLDSDMDODMDODMDMPPDMDMDODMDSDMDMDOOTDORMDMDODMDMDODMDMDMDMDMDMDSDMDMPEDMDOOLNLDMOTDOOLDMPMDMRMDMDOPPOTDMDMDMRMDMRMDSDODMDMDSPPDMDSPPDMDSDMDMRMDMDMPERMDODMDMDMDMDMDODMDMDMDSDODMDMRMDSRMDMOLPPRMDMDSDODMDMDODSDMDSDMDMDSDSDODMDODMNLDMOLDMPMOTDODMDMDMDODMOLDMOLDMNLPPDMDMPEPPDSDMDSDMDODMDSDMDSDSNLDMDMDMDMDSDMDMDMDSNLDMDMDMDMDMRMOLDMPPOLDMDMNLDMNLDMDMDODMDMOTNLDODMPPDMDMPPDMDMDODMDSDSDSDSDODODODMDMDMPPDSDMDSDMNLDMDMDMDMDMPPPPOLDMDMDMDMOTDMDMDMDMNLDSNLDMDODMDMOTDODSDOPEDMDMDSDMDMPEDODSDMDSRMDMPPPPDSDMDMDMDOPEDMPPDMDMOLRMDMDMOTDMNLDMOTDMDMDMDSNLDSDORMDMDODSDODODMOLDSDODODMOTDSDMDSNLPEDSDMDMOLOLDMDMDMDSNLDMDOOTOTDMDMDMDMDMRMOTDMDMOTDMNLDMDMDMDSDMDMNLDODMNLPPDMPEOTDODSDSRMDSOLDMDSDMRMRMDMDMDMRMDMDMDMOLOTDSOTDMPEDMDOPEDMDMDMDMRMRMDMDMRMOTOTDMRMDMDMPENLOTRMRMDMDMOTOLNLDMDSDMRMDMPERMDMDORMDSRMDMDODOPEDMDMDMDMPEPPDMDSDODMDMNLDMDMDODORMNLDMRMRMDMDODMOLPEDMDMDMDMOTDOOLNLDMDMDMRMDMDSRMDSNLDMDMDORMDMDODSDMDSDMRMDMDSPEPEDMRMDMDODMDMDMDONLOLNLDMDMPEDSDOPMDMDMDSNLOLDSDMDODMDSRMDSDMDSDMPPDSDODMDMDMDMNLDMPENLPEDMDMNLDMOTPMDMDMRMPPPMRMDMOLOLNLNLDMDMRMDMPMDSDMRMNLNLDSDMDMDSDSPPDMDSDSDMDSDODMOTDMDMRMRMNLDMPMDMNLDMDMDODODMOTNLOLDMDMNLNLPEOTDMPMNLPMDODMNLDMDMNLPPDOPPDODSRMDMDODMPEDMDMNLDSDMOLDMDOPPDMPPDMRMRMPMDMDMNLNLDMDMDOPFNLDMNLDMPPDMPMDMDMDMDODMDODONLDMRMDMRMOLDMNLDMDSPPOLDMDSDMDMDMDMPPPPOLDMDMDMPPNLDODMDMNLDMDMDODMDMPPDODMPPDMPPDSDSDMDMDMDMDMDMDMDMDMPPDMPFDMDODMDMDMDOOLDODMPPNLDMDMNLPPDMDMDMOLPPDMDSDMDODMDSNLPPDMDMDMDMDMDMDMPMDODMNLDMDMDMOTDMDSDMDMNLDMNLOTDMDODSDMPEDMDMDMPEOLDSDMDMOLDMPMDODMDOOLDOPMDMDMDMDMDMDODMDMOTDMDOOLDMDMDMDSDOOLDODSDSOLPMDMDMDMPMPMDODMDORMOLDODSDSRMDMDMDMDMDMDODMPERMDODMDMDMPMDMDODODMOLRMDODODMDMNLPEDODODSDSDSDMDMRMDMRMDODSDMPMDMRMPEDMRMDMOLDMNLRMRMRMRMRMDONLDODMPMDODMRMNLNLDMDODODMRMRMDOOTDMPMPEDMDMOLRMRMDOPMDSDMDSDSDSDMPEDODORMDMDODSDOPEPMDMPEPMDMRMRMRMRMDORMRMDMPMRMDMDOOLOLDODMDMRMRMDMDONLPMDODONLDMDSDORMDMDMPMDOOLDODSDODODMDSRMRMNLDMDMDSDSRMRMPMPEDOPERMOLRMDODMRMRMDMDMNLDODMOTDMDODOPMPMDMPMDOOTNLDONLDODMRMPEDMDODODMDMDSRMDODONLRMDODSDMDSDOPMDSDSDMPEPMPEDMOLDMPENLDMOLPEDODORMPPDMPEPMOLPENLDMDODMDMDMDODODMNLPMNLOLPEPEPPDSNLPPDODSRMDSNLDSDONLDMDSDSPPPMPEDMDMDMDMOLNLDMPMDOPMDOPENLNLNLDMPMPERMDSDMPMDMRMPMRMDMDONLPEPMDSDOPEOLDMDSNLDSPEDMDMPMOLDMRMRMDMPERMDMDMNLDMPERMDMDMDMNLPMDSPMPEDMRMDORMDOPPDMRMDMRMDMRMNLNLRMOTDMRMDOPEDOPENLDMPMDMDMPMPPOTOTNLOLDSDMPPPMDMDSRMRMRMDONLDMRMDSOLDSDMDOPEDMDMDSDMPMRMRMRMNLDMDOPMPPDMDMDMDMPERMRMRMOTPMDMDODMDODODMDODSDMDMRMDMRMOLNLNLDMPEPMPPRMPEDMPEPEPMRMRMRMOLDMOTDMPEDMDSDMRMRMPMDMDMNLDSPMDMDMPPDODMPEPPDSRMPMPPDMNLRMDMOLPPNLDMDOPERMPEDMDMDORMPEPEDMDMOTRMDSNLNLOLPPRMDMNLDOPPDMSHDMDSNLRMNLOLDMDONLPMPPOTDORMPPDSDMNLDMDSDMDMPPPEPEPMDMPPRMDMDONLOLPERMDSNLDMOLDONLNLDMDMDMDMDMOTNLDMDMDMNLOLDSPPOTPPPEDMDMDMOTPEDMPPDMOTOTPPNLDODSDSPEPMPEDSNLNLOTOTPPNLDMPEDMDMOLPMSHNLPEOLDMDMOLDMDMDODMPEDONLRMOLDMRMDOPMDODSRMRMRMPMRMDMNLPMPEPEDMPENLDMOLDMNLPPPEDOPEPMDSDSSHDMPMDMDODMNLDMDMRMOTNLPFOTDODODMOLDODMPEDMNLDMDMRMDMPEOLPEDONLDSRMDMPMRMDODMPERMRMRMOLRMRMPMDMDMNLRMRMDMNLDOOLPEDMPEDOOLRMDOPEOTRMPMDMPMDMPEDMRMPENLPMDSDMPEDMNLRMDMDMRMPFDODMRMNLDMOLPEDMDMDMRMDMPEOTSHRMDORMDODODMRMRMRMDMRMDMDSRMRMDONLPEPMPERMRMDMNLPEPEDODMRMDOSHPMDMNLRMDMDORMDSRMPERMDODMNLPMOLDMPEDMDMDORMDMPMDODMRMRMRMDORMRMDOOLPEDMDONLDSDORFRMPERMPMRMDMPEDMOTSHPEPESHDMDMDORMDSNLRMNLOTPEDODMSHPEPMDODMRMPERMDORMRMDMPMRMRMDODORMOLNLRMOLDMDMRMRMDMPEDMDMPMDODODORMPEDMSHRMDORMDMOLDSOLNLPEDODMRMOTPEDMRFDOSHDSDMDMDOPEDMDMPMDMPEOTRMRMDMPEDMRMRMNLRMRMRMPMOLDODMDMDMPMDORMDMOTPEPENLDMPEDSRMRFRMOTRMDOPEOTPMPEDSRMNLPMDMNLDMDOOTOTSHPEDODMOTOLNLDMDMOLOLPEPFOLDODMDMDMPENLDOSHDMDODSDSOTPPPEDSOTPENLPEDMSHDMDMNLDMPEDMNLDODORMPPDOPEDMPMOTPENLDMDSDMDMDOPENLNLOTDOPMDSRMNLRMPERMPEPEDSOTPPPEDOSHPENLPEPPOLDMOLRMRFDMDODMPPNLRMPPOTDMPENLDMDSDMDMNLDMPEDMPPDMPENLOTNLDOPEPPOLNLSHPEPPDSPMDOPEPPRMDMPPDMNLPEDODMDMDODMDMPPNLOLDMDMDSDMOTPMDONLOTRMNLDONLPEPEDOOLRMOTPEDSNLPPDMPPPPPPNLRMPMPPSHPEPPPPNLDMDMNLDMDODMPPPERMPEDMPPPEPEOLDMDSDMDMDMPPDOPPDMRMSHDMNLOTNLPERFDMPPDSDONLDMOTDMPEPEDMPPDODMPEPEOTOLDMDMRMOTNLDMDMDMPEDMPEPEPEPEPESHDOOTOTDMNLNLOTPEDOPPOTDMDMDODMRMDMNLDMOTDMDMPEDMDSOTDMOLPEPEPENLSHPEDOOLPERMDMSHRMOTNLPMRMDMPEPERMPEOTSHPEDSOLSHNLSHPEOTDMOLDMDMOLRMDMRMRMDMPEPEOLDMOTPEPERFDOOLDMOLOLDMPEDSDMPENLOTRFDMNLRMPERMRMOLRMNLOLRMPEPERMDORMRFRMDSRMDMDMPEDMPEPEOTRMSHRMRMRMOLDMDMOTRMDMOTDMPERMDMOLDMRMRMDORMDMPEOTSHRFOTDMOTOLRFPEPERMDMNLPMRMRMNLRFSHSHDOOTDOOTRMPERMRMSHOLNLPERMDMDORMPEDMRMDMRMRMRMDMPERMOLDMPEDOOLRMRMRMPEOTRFOLDMRMDMDMRMPERMDORFRMPEOLSHNLRFRMNLPEDORFDMOLPEPERMPMDSRMDMOTDOOTPMRMRMRMRFRMSHSHRMRMRMPEPERFRMRMPENLRMRMRMDMDMDMRMRMOTPEPEDMPERMDMRMOLRMOTRMPEDODMOLRMRMDMRMDMOLRMOLNLPERMOTNLRMPEOLRMRMDSRMRMPEPEDMRMRMDSPERFPERMRFOLDORMRMPERMSHOTOLRMRMRMNLPEOTDMPESHRFPERMRMSHSHDMOLRFDMRMRMDMRMPERMDMDMRMRMPMRMDMSHPEDONLOLRMRMSHDMRMDMPEDORMRMDMOTOLRMDMOLRMRFDMRFDORMPPRFSHDMPERFNLPESHRMRMRFOTRMRMNLRMDOSHNLRMPERMRMRMPERMDSDMRMDMRMNLPERMPEDOOTNLSHNLDMRFDORMDMOTDSDOSHPMDODMRMPENLPEPENLOTDOPMDOPMNLSHPPRMDMSHDMRMDMDMDMPERMPERMOTPEDMRFRMRMDMDMSHRMDMRMDMRMOTNLDORMNLPENLSHRMDORMPEDMRFNLRMPMRMRFDOSHPMRMRMPEOTNLSHDSRMRMSHRFSHDODMRMDMPERMDMPEDMRMOLDMDMRMPEDOSHNLRMNLPMRMRMRFSHRFRMRMPEDMPMNLDODSNLRFPERMSHNLRMDMDMSHDORMRMRMDMPEDODMRMDMDMRMDMOLRMRFRMDMRMOTOTPPDMSHRMDODSRMDMRMDODOPMPPDODODMDMDMOTRMDODOOLDMDMDMRFDMRMPPPEDMPERMNLRMDODMDMDMDMDOPPDOPFSHDOOTDODMOTDMOTDSOTPENLPPDOOTPESHDODOPEDMPPDMPPDODMSHOTDMDMDMDMDMOTDODOOTDMOLPEDMPPSHDSDMDMRFDORMRMDMSHDMRFOTPPDOPERMNLRMPEDMDMOTPFPFDODMNLRMDMPERMRMPEPFOTRMPENLRFRMRMNLRFOTPHRMPFDSRMDORMRMOTNLOTDODORFPERMDOPERMDOPFDORMSFOTOTDMPEOLOTDMDMOLDMRMDMOTRFPFOTPERMRMDMOTRMDODMDMRMRMDOOTSFRMRFRMRMPEPERMDORMRMRMDSPFDODORMRMDORMRMPHRMRMRMNLRMRFRMRMRMRFRMRMDOPEPEDMRMSFRMDMRMDMPEDORMDMRMDORMNLRMDMRMRMDMPEDMDMRMDMRMDMRMDODORFRMDMDMRMSHDOOTOTSFSFRMRMRMPFNLDODOPERFDMDMDMBADOPEDMOTDMDODODMDMDMDMDMRMRMRMDMNLRMPESFRMPFRFRMDMRMRFSFPEPFPHRMRMOLRMRMPEDORMDODMDMRMOTRMPEDORMOTDMDMDMRMDMDMPEDMRMDORMRMDMDMDMRMRFDMRFDMDMDMPFRMRMRMDOSFPFRMNLNLRMRMRMOLPHPERMNLPESHOTDODORMDODMDMBARFRMRMDMDMDMDMRMDODMDODMDMRMDMDMDMDMRMDMRMDMDMPEPERMDMRFRMDMRMPFPERMPERMDORMPHNLOTNLRMOTRMPFRMRMRMRMPEDMRMRFDMDMPFRFDMDMRMOTDORMDMDMRMDORMRMDOPFDMDODMDMSHDMDODMDMDMDMRMPEPFRMRFRMDMRMRMOLPHRMPEOTPPPEPEDOOLDMDOPEDONLSHDMPPOLPPDMDMDMDMDMRMDMDODMRMRMDMRMDMDMDODMDODMDMRMRMDMDMPFOTRMRMNLPEPHPFRMSFDMDODOOTDMRMDMDMNLDMRMDODODODMOTDMDMDMSHDMRMDMDMOTPPBAPFOTRMDMRMRMOTDOSFOTRMPPNLPERMDMDMRMDMBADODMDOPFDMDMOTDMDMRMDMDMDMDOPPDMDMOTDMPEDMDOOLDMNLDOOTOTPPRMPPPPPHDMPHPPDMRMDMDMDMDOOTRMDMDMDMDMDMOLDMDMDMDMDMDMDMNLPPOTDMRMPPDMDODODSDMDMDMDOPEPFOTBADODMOTDMDMDMDMRMPFPEDMDMDMDMDMDMDMPERMPPPEPFDMPPDONLDSDOPPOLDMDMNLDOPEDODMDMOTDOPEDMDMDMDMDMPPDONLDMDMDMDOPFRMRMRMOTPFOLDSDMDMDMDMOTDODMDMDOBADMDMDODMNLPPDMDMDMDMNLDMDMDMOTRMOTOTRMDMOTDMOTDMDMDONLRMPFRMRMDOPERMDSOTDMDORMDMRMNLDODMPEDMOTDMRMDMRMDMPFRMDMOLDMDMDMDMRMDMDMDMDMDODOPFRMDMRMBADODMPEDSOTRMDMPEDORMRMDMOLDMDONLDMDORMDMOTDMBAOTPFRMPEDMRMRMRMDMRMDMDMDMPEDMDMRMDMRMOTOLDMDMDMDMDMDMRMRMDMDOOLPFRMDMRMDOOLRMRMDORMDODMOTOTRMDSRMRMNLOTRMDODOPEBARMDMRMDMDMRMDMDMOTDMRMDMDMPPDMRMDMDMDMRMRMDMDMDORMBARMDOOLDMOTRMDMDMDSNLOTDORMRMDORMRMDONLOTRMRMRMDOPEDORMRMDOOTDMRMRMDMRMRMDMDMDMRMDMDMOTDMDMOLDMDMRMDORMDORMDMNLDORMDMDODSRMDODMDMRMOLDONLPERMRMDORMRMDMBADMDMRMRMDMDMDMDMDMRMRMDMRMRMDMPEOLRMDMRMPPDODMNLBANLRMDONLPPDORMRMRMDMOTDMBAOLPERMDMDODMDMDMOLDODONLDMDMDMDMDMOTRMDMRMDSPPDOOTBADMDORMDODONLPENLRMOTDODMBAPPOLOTRMDMRMDMOLDMDMPEDMOTPFPPPPPPDMDMPPDMOTOLRMPEDMPPRMRMOLPEDSDOPEBARMDOBAPPDMDMDODMOLDMPEDMDMDMDMDMDMDMRMOTPEDMPPRMPPBARMDMDSDOOTPPDONLDOBAOTPPOTPPPPDMROPPPEDMPFRMPPPPDMPPPPPPSFPPDMDMDMOTRMPEPFPPPPPFDSOTSFRMNLBASFDODMDOPPDMDMPPDMPPPPPPOTDMRMDMPPDMOTRMPPDMPPDMOTPPDMPPOTOTDMRMDMPPRMDOOTBAOTOTPPDOPEPPDSDMPFDOPPOTDMDMSHPPPFPPOTDODMDMOTDMDMDMPPDMOTPPDMPPPEPFPFPPPFDMRMPPPFPFDOPPBAPEDMPFDSPFDMRMPFSOPPDODODOPPDODMPEPPOTPFOTDMPEDMPPPPPPPFDMDMDMOTDMPFOTPPPPDMPFPPPPDMOTPFDMPFPFPEDMRMRMPEOTDOPFOTRMRMOLSOBAOTRMRMDMPFDSOTRMRMRMDMRMDMNLPFRMPEDODMBADMRMNLDMPFSORMBAPEPPPFRMSOSODORMRMPEDORMRMRMRMSORMDOPPDMDMDMRMRMDMDMDMDMPFPFDMOTPFRMRMDMPFRMRMRMRMPFDMRMOTOTBARMRMDOPERMRMRMRMDORMOLDORMSODOPFDODODMPEPERMRMRMOTPEPERMDODMRMDMPPRMRMDODMDMSOOLPFDMOLSORMBAPEPFRMOTDMDMPFPFDMNLRMDMRMDMDORMRMDMPFRMDMRMDMDORMRMBARMDORMDMDORMDMPFOTDODMDOPPRMPERMDOBAPEDMPHNLPEOTRMDMDMDODMRMRMOTRMSOOTDOOLRMOLRMRMPFRMSODOPEDSDORMDMPEPEDMRMPENLDMDMSOPERMDMPPDMDORMDODOOTDODODMDODMDMPESOSOPERMRMPHDMRMPFRMBADOPPPFRMPEDMRMRMBADMPEPEPFDOOLRMRMRMDSRMPEDORMDMPPRMDOPERMDMDODMRMPESORMDODMRMDMDMPHRMRMRMDODMOLPFRMOLDODSDSPERMDOPEDODMDMPHDOPFDMPFRMPEDMDMDODMDMRMDMPPDMPPDMDMPPDODMPPPEDMDMRMOTPPDSSODODMPPPHRMSODMOLSOPPDMDODMDMDMDMDMDMPPDMDMPEDMOTRMOTDMDSPPDMDODMDMNLPPDORMPFDODMNLPPNLDMPFDMDMDMDMRMDMOLPPDMDMDOPENLRMPPDODSPFPPDODODOOTRMRMRMDMPPDMDODMOTDODMRMPPPPPFPPOLPPRMDMDMDMDMSOOLDMDMPPOLPPSOPPDMPPRMRMDOPPDORMPPNLPPPPDSOTRMDMDMRMPPOTPFDODMDMRMDMDMPPDMDMDMSOPFPPPFPFDOPPPPPFRMPPPPSOPPDOPPPPDSDMRMDMOLDOPPPFOLPFDMPPPPPPRMDMPPDMDMDMDOPFDMPFRMPPPPDSDOPPOLPPNLPPDMDOOLRMRMPEPPRMRMSOOLPPDMPFPPDMDMDMPPDMOLDOPPDOPFOTPFDMOLPFPFRMOLDMRMNLPFDMSODMRMOLRMRMDMDMRMPFPFDMPFPPPPDMDMDMDMRMDMDMDMRMRMRMPFRMRMOLDSDMRMDMDMDMRMRMSFDMDMDMPFRMRMDMRMDOOLPFRMRMOLDMDSOLDMRMDMDMDMRMDMRMOLRMRMOLDMPFRMDMRMDMRMDORMRMOLDMDMDMRMDMOTRMSFDMDMDMPFDSDMPFPFDORMRMDMOLRMNLPFDMRMDMDMDMDMDMDMDMOLRMDMDMDMDOPPPPPEPFRMPPPFDMPPPPPPPPPPDSNLNLRMOLPPDOPPDMSOPPDMDMDMDMDMRMDMOLDSOTPPDSDOPFPPDMDMPPDOOTOLDODMDMDMRMDMDMPEPEPPOTPEDOPFDMPPDMPPRMDSPPPPPPDSPPPPPPPPDMPPDMPPDMDMDMPPPPOLOLPPPPPFPFDOOLPPPPPPPPPFDSPPPPPPPEPPNLRMPPDMSODMRMPFDMPFDMPPOLPPOLPEPFDMPPPPRMRMPFOLDMPPPPPPDMPPRMDMDMDMDMPPDMPFDODMPFPFDMPPPFPFDMPPPPDMPPPFPPDMRMDMRMPFDMDMRMPFDMOTPFDMPPDMPPDMDMDMPPDMPFPFPEPFOTDMDMPFSODMDODMDMPFNLDMPFPFRMDMRMPFDMDODMRMDMDMPFDORMDMRMOLDMDMOTRMDMDMDMRMDMSOSOPFDMOTDMOTDMDMDMDMDMPFPFDMDMRMPFDMPFDMRMDMRMDMDMPFRMOTRMDMDMDMDMRMDMPFDSDMDMOTPFPPDMDMDMPFPPDMDMRMDMDMDMDODMDMRMRMDMPPPFDSPPPPDMDMPFDMDMDMOLPFDMPPDSDMPPDMPPPPOLPFPPPPDMOLDMPPDMDMDMDMPFPPDMPFPPPPDMRMPPDMPPDODMOTRMDMDMPPRMDMPFPPPFPPPFDMPPPPPPDMPPOLPFDMDMDMDMOLRMDMPFDMOLPPDMPPDMPFPFPPPPRMPPPFPPRMOLDMDMRMPPPFDMPFOLDMPFDMPPDMPFPPDMPFPFDMRMDMPPDMPPDOPFDODMPPPPRMPFOLDMDMPFRMDMDMPFPFDMPFDMDMPFDMPFDMDMPFPFDMPFPFPFDODMDMOLDMDMPFPFPEPFDMPFDMDMDMDMDMPFDMDOPFDMPFRMOLDMDMDMPFPFPFDMPFDMDMDMDMRMDMDMOLDMDODMDMDMRMDMDODMPFPFOLDMRMRMDMDMDMDMDMPFRMDMOLDMDMDMDMRMDMPFDMDMDMDMDMPFPFDODMPFRMDMDMPFDMRMDMDORMOLDMRMDMDMDMDMRMDOPFDMPFPFDMDMRMDMPFPFDMDOPFDMDMDMDODMDMDMDMDMDMDMRMRMDMPPPERMDMDMDOPPDMPPOTDMDMDODMDMDMDMDMDMDMRMRMDMDMDMPPDMDMDMOLPFDMPPDOPEPPPPPMDMDMRMPPDMDMDMPFDMDMPPDMDMDMDMPFOLDODMPFDMPPDMPPDMRMPPRMPPPFPPDMOLPMDMDMDMPFPPDMDMDMDMPPDMOTRMDMPPDMOLDMDMPPPFPPPPDMDMPPPMPPPPRMPHDMPPDMPPPPPPPPPFPFDODMPPNLPPPFDOPFOTDMDMOLDMDMRMPPPPPPDMPPDMPPPPPPPFOLDMDMRMPPPPPPPPPPPFDMPPPPDMOTDMDMPPPPOTPPPPDMPPPPPPPFPPPPPPOLPPPPRMPPPPRMPPDMDMPFPPPPDMPPPPDMPPPPPPDMPPPPPPPPPEPPPPOTPPDMPPOTDMPPPFRMRMPPRMPPPPPPPPPFPHPFPPPPPFPPPFRMDMPPPFPPPPRMPPPFDMDMPBDMPPPFPPOTPPPPPFPPNLPFPPDMPPPPPPPFPPPFDOPEPFPPPPDMPPPFPPPPPPDMPPDMDOPPPPDMPPOTPPPFOTPPOLPPPFPFPPDMPPDMPPPFPPPPPPPPPPDMPFPPPEDMDMPPDMPPPFPPPPDMOTPFPFPPPPPBPPPBPPPPPPPPPPDMPFPPPFPPPFOTPPOTDMNLPPPFPEPPPFOTPPDMDMPPPFOTRMOTDMPPPPPFPPPFDMPBPPPPDMPPPPDMPPPPPPPFDMDMPPPPPPPFPPPPPPDMDMPFPFPFPPRMDMPPRMPBPPPFPFDMDMPMRMPFDMPMDMDMPPPFDMPPPPNLPPPFPFOTOTPFRMPFDMPPPPPEDOPFDMRMDMPFDMOLPPDMOLDOPPDMOTDMPPDMRMDMDMDMDMRMPPNLPBDMDMDMDMPBPEPLOTDMDMDMPFDMPFPFOTPPPFDOOTPFRMOTDMOTDMDMDMPFPFRMDODODMDMDMPPDMPPDOPFPLDMPFDMPBDMPPPFDMPBDMDMPEPMDMPLDOPEDODMPFDOPFPFDMPFPFPFPPDOPFDMDMDMDMDMDODODMDMPFDMDODOPFDMPLDMDMDMDMDMPPRMDMPPDMDMDMPBPFPBPEPFDMDMDODMPMOTPEPFPEDMPFPMRMPPPEDOPFDOPFDMDODOOLOTPFDOPFPFPFDMDODMDMDMPFPFPFPFDOPFPPDMPFDMDMPFDMDMPFDMPFOTDODMDMDMPPDMDMPFRMPPDMRMDMPFRMRMPPPFPEDMOTOTPPRMPFPFPMPPDMOTPEDMPFPFPPPPPPRMPEOTPFDMPFPFPPPMDMRMDMDODODOPMDMPPPPDODMDMDMPPPMDMPFPFPFDMPFDMPPPFPPRMDMPFPPPFRMDOPPPFPPDMDMDMPPOTRMRMPEPPPFPEDMPEPFDODMDOPPDMDOOTPMPFDOPFDODOPPDMDMPFPFPPPFPFPMPPRMPPPFDMDOOLPPPFPFDMDORMPPPPDODMPEPFRMPFDOPFPPPPDMDMPFPPDOPPDOPPDMPPDMDMPFDMDMPPPPPFDMPPPFDMDMDMPFPFPPDMPPPPPEDMDOPPPFDMDOPFDOPFPFPPPFDOPFOTDMPFPFPFPPPFPFDMDMOTDMDMPBPPDMPFPPPFPPDOPPDOPFPPPPDMPPRMPFPFOTPFDMPPPPDMPPPFPPPPDMPFPFDMDOPEPPDMDMDODOPFPPPPPEDMPFPPPPPPDOPPPFDOPPPFDMDMDOPFOTDOOLPFPPOTPFPPDMPPPPPPPPPPPPPPPPDMPPPPPFPPDMDOPBPFPFPFPPPPPFPPDOPPPFDODOPPDMPPPFPFDODOPFDMPFDMDODMPBDMPPPPPPPPDMPPPPDMPPPBPPPPPPPPPPDOPPPPPPDMPFPPPPPPPPDOPPDMDMPFDODOPFPFPPOTPPPPPFPPPFDMPPPPPFOTDMPPDMPBOTPPDMDMPFPPDMOTPPOTDMDMPFDODMPPPPDMPPPBOTPPPFPFPPPPOTDMPPPFPPPFPFOTDMOTPFOTPFOTPBOTDOPFPPPPPPDMPPPPPFOTDMDMPFOTPFDOPFPFOTPBPPOTDMPPDMOTRMPFDMPFPEDMPBPFDMDMPPDMDMDOPFPPPFDMPLDMDODMOTDMOTPFPFOTOTPPDMOTOTDOPBDMPPOTDMDOPFPFPPDOPFOTPFOTOLOTDMPFPFDMDMDMPFDMPFDMOTOTNLPBDMOTDMDODMOTNLDMRMPEPMDODMPBPFPFPFDMDMDMRMPFPBPPOTDODOOTRMDMOTPFDMDMPFOTDMDMPFDMDMDMPFDMDOPFOTDOOTDMPFRMDMDMOTPMRMOTDMOTRMPMPFPFPFDMDMDMDODODMPMDOOTPFDMRMRMRMPBRMPBDMPMPFRMDMPMDODOPMDMDMDODMOTDODOPBDMOTDMPFRMOTDMOTOTRMDMDODMDMPMDMOTDMDMPBDODMRMPFDORMPEPMRMDMRMDMDMDODMDMRMPFPMPLPXDMPBPMRMPMDMDMPFDODMDMRMPBDMDMRMOLDODORMDMDMRMDOOTOTRMDODODOOTDMPFPBDMDORMRMOTPLDMDMDMDMDOOTPMDMDMOTRMDMDMOTPFDMPMDMPFRMPFDMRMPFOTRMRMPMRMDMRMRMDMDMRMDMDMPFPFOTDMRMDOPFPMDORMDMPMPEOTDMPMOTRMOTPFDODMDMDODMDORMDOOTDOOLDORMRMOTPEOTDMDMOTDMDMPFDMOTPBDMOTOTDOPMDMDMDORMOTPFDORMDMPFDMDOOTDMDMDMDMDMPMDMPFRMPMPMPPPPRMRMDMPFPMPPPPOTPPPMRMPPDMPEDMPPRMPPPMRMPPPPDMPFPPPMRMPPPPPMPFDMDOPPOTOLDMOTPBPEDMPPPPOTDMDOPFPMPFPPDMPPDOPMDMPPPMOTDMRMRMPPDOPMOTPBDODMPPPPDODMPPOLPFRMPPOTDMOTDMRFDOPEPMDMPPDOPMPMDMDMDOPMPFPFPPDMPPPBOTOTRMPPOTDMPFPPDOPFPPDMOTRMPFDMPMPPPMOTPMOTOTDMPPPBNLSOPFOTPMPMDMPMPFNLPMOTPPPPPPDMPPPPRMDMPMPPPMPMPPOTDONLOTPMPMRMDMPFDODOPPDMPFRMOTPMDOSONLPFPPDMPMPEDMDOPPPPPPPMDOPFSOPMPMPMDMPERMDOPPOTOTOTDOOTOLDMPFPBDODOPMDMPPPPPPRMPMDMPBPBDOOTDMDODMOTDMDODMPPPMPPDMDMOTPBOTPPOTOTPBOTOTDMNLPBPPPPPEPBPPDOPBPBOTPBPFOTPMPFPMOTDOPBDOPMPPPPPMPPDMPBOTDOPFPMPBDOOTPPPBOTRMPMPPOTDMPMPPDMDMNLPPPMPMNLPPPPDMPFPPPMOTOTPPDMPBPMOTDMPMPPPMDMOTPPOTPPDMDMDMDODMDMDODMPPPPPMPPOTPPPFPBPMPMPPPMPBDMPPPPPMPBPEPBDMPBNLPPPBPBPMDMPMPFPPPPPMOTDMDMPPPBPBPPOTPMPPPPDMPFPPDMDMOTPPPPPMPPDOPFPPPPDOPPDODMPBPFPMDMNLPEPPPBPPPPPPPPPMPPDMPPPFPMPPPPDSPFDMDMPBPBOTPMOTPBDONLPPOTDMPPPBPPPPPPNLPMPFPLPMPBPMDMDMDMPPPPPPPPDOPEDMDMDOPFPBPPPBPFPMDMPPPPPFPBOTPPPBPPNLPPPBPLPPOTPPPPPPPPPPPMPPDMPPDOPPPBPPPBPMPFPFPPDMSHPPDMPPRMPBOTPPPBPMOTPPPBDMPFOTPPPPPPPPDMOLPPOTDMPBPPPBDMPPPFPFNLPBDMPLPPDOPEPPOTDSPPPEPFPFPPPMRMNLOTDMPPPBPFDMPPPPDOPMPPPBSHNLOTPPPPPFPPDMOTPPSHPPNLDMPPPPDONLPPOTRMPPDMDOPBOTPPDODOPFPMPPPMPPPFDOPBPBPMPFOTPPDMPMPPPFDMDMDMPBDMPPPPDOPBPPPFDMPMPPOTPMDMNLOTPMPLPPPPPPDSNLDMPBPFPFPMDMDMOTDMPEDOOTPBPBPBOTOTPBNLPBPMDOOTOTDMDMRMNLPBOTPMPBDMDMDMOTPFOTPFPLDMPBPBPBDMPMPBPBOTPBDOOTPMOTOTPLPBPPPMPPPBPMPBPMDODODMPBDMDMPBPMPMPFNLOTPMDSRMDMPEPPNLOTPEPBDMPBDMPFOTPFPMDOPBDONLPBDMOTDMPMRMDMPMNLDMOTNLPBPBDOOTOTDMPBDOPEDOPMPMOTDMDMDODMPBDMDMPBPMPMDMPBPMDOOTPMDMRMPBPMDMDMOTPMDODMPBPMPMDOPEPBNLDMPMPBDSOTDMPEPMRMNLPBPMPLOTOTDMPBDMDMDMPBPBDOOTDMPMDMPEPMOTDOPBDMDMPMOTSHDMDMOTPMOTOTDMOTDMPMDMDODMDORMOTDMDMPBNLDOPBOTPBDMDMPBPBRMDMDSPBNLPMPMDMDMPBPMPMDMNLRMRMOTDMDOPBDMDMOTPBPMDOPMOTPBOTDOOTDMPMDODMDMNLPBRMOTDMDMDMOTDMDMDMDMDMDOPBOTDODOPMOTDMDMPMPMDSDMDMDMDMDMOTDONLPBDMDMOTDMRMOTDMPBPMDMDOOTPBDMPBDMOTDOPBDMDOPBDMOTDOOTDMDMOTDMDMDMPMPBDMPMDMOTDMDOPMPLDMPMDMPBRMNLDMOTPMPEDMDMDMPBDMPBPBRMOTDOOTPBPBPBDMDMPMDMOTOTDODMPBDMDMDMPMDONLPBDMDMDOPMDMDMDMDMDMPMDMDMDMOTOTDMPMOTPBDMDMDMDMDMPBDMPBDMPBDMPMDODMDMOTDMDMPBPBPEDMDMDMNLDMDMDMPBOTPBDMPEDMNLDMPMPMNLPPDMDMDMPFPBDMOTDMDMPBPPOTPBPPPLRMPPPMPPOTPBPBDMNLDMDMPPDMPPPMDMPPDMPPOTDMPPDOPPPPDMDMDMDOOTDMPENLOTNLPBDMPPDMDMPMDMPFPFDMDMDMPBPBDMDMDMPPPBDMPPPMPPDMNLPBDMPFPBPBNLPMDMDMDOPPDOPBPMPPDMOTPPPPDMDMPBDMOTDMDODMPBPBDMPPDMDMPMPMPPDMOTPBDMDMDMDODODSPPDMPPNLDOPPPPDMPPOTPBPBPFNLOTPBPBPBOTPBPFPPPEPBPFDMDMPPPBPPDODOPBDMPFDMPBDMPPPPDMPEDOPBPPPBPPPMNLDMDMDMPBDMDMPBPFDMDMPBPPPBPBPMPMOTPPPBDMDMPBPBPMPBDSPPPFPPDMPPDMPFPPDMDMPBPPPBPBPBPPPPPPPFPBPPPBOTOTPBPBPBPFDMPBDMPEPPPBDMPBPBPBPPOTPPPFDMPPPBPBPPPPPPPBPPPBOTPPDMOTNLPBDOPBDMPMPPDMPPPBPMPPDMDMDMPPDMPPPBPPDMPPPBPPPBPMPPPMPPOTPBPBDMPBRMPPPPPPPBNLPBPPPBPBPPDMNLOTDMDMPPDMPPDMPPPBPPPPPMPBPPPPPPPFPFPBOTPBPBPBDMPBDMPBPEPPPBPBPPPBPBDMDMPPPMPEOTPBPPPPDMPPPPDMDMPPPBPBPBPPPBDMDMOTPMPPPPPBPBNLPPDMDMPBPPPBPPPFPPPBPBPBDMPEDMDODMPPPPDODSPBPBPBPBPPDMPPPMPMPMPMDODODMOTPBPBPBPPOTDMPPDMOTPPPMDMDMPPDMPPPMPBOTPBPMDMPMDMOTDMDMDMOTPBOTPBPBPBDMPBDMOTPMPMOTPBOTPBPBDMDMPMPMOTDMDOPBDMDMDMDMPBPBPBOTDMDMOTDOOTDMDSOTPBDMDODMDODMDMDMDMOTPBPBPBPBPBPBRMPBPBPBPBDMPBPMPBPBPBPBDMDMDMOTPEOTOTOTDMDMDMDMOTPBOTDMDMPMDMDOOTOTDMDSNLDMDMDMDOPBOTPBPBPBOTOTDMDMDMDMPMPBPBPBOTDMPBPBPBPBPBPBDMOTDMDMDMOTDMDMOTPBPBDMPBDMOTDMDMDMDODODOOTDSNLDMDMDMPBPBPBPBPBPBOTPBPMDMDMDMDMDMPBOTDMPBPBPBPBPBOTPMPBPBPBDMDMDMDMOTOTOTDODMOTDODODMDMDOPBPBDMDMPBPBOTDMDMPMDMDMPEDMDMDMDODODMDODSDOPMNLDMDMDMPBDMPBPBPBPBPBPBRMPBPBDMDMDMOTPMRMPBPBPBPBDMPBDMPMPBPBOTDMDMDMDMDMOTOTDMDODMDODMOTDMDODMPBPBPBPBDMOTDMDMDMPEDOOTDMDMDMRMPMNLDODSPBDMDODMNLOTDMPBPBPBPBPBPBDMOTDMDMRMPMRMPBPBPBPBDMDMPBPMPBOTPBDMDMDMDMOTOTDMOTDMDMDMDMDMPBOTPBPBOTDMDMPMDMDMDODODMDSDOPPDMOTNLDMPBPBPBPBPBPBPMPBDMDMDMOTDMPBPBDMPBPBPBDMDMPPPMPBPBPBPBPBPPPBDMDMDMDMPPOTOTDMDMDMDMPBDOOTDMPBPBPBOTDMDMDMPBPMDMPPDMDMDMPMPBNLPBDMPPDMOTDMPBPBOTPPPBPBPPPBDMPPDMDMDMPBPBPBDMDMDMOTPBPBOTPBPBPBPBPPPBPPPBOTDMOTDMDMPBPPDOOTPBOTPBPBDMDMPPPPPPPEPPOTDMDMDOPPDMPBOTDMDMDMPBNLPBPBPBPBPBPBPBPBPPPBPBPBPBPBDMDMPPDMDMPPPPPBDMPBPBDMPBPPPBPBPBPBPBPBDMOTDMPBPMOTDMDMDMPPDOPBPBDMPPOTPPPPPPPPNLPPNLPBPBPPPPNLPPPBNLPBPBPPPPOTPBPPPPPPPPPPPPPBNLDMDMPPPPPBPBPBPBPBPBPBPPDOPBPBOTPBPEPBPPPBDMOTDMDMPPDOPPOTPBPPPPPPPPPEPPPBPPPPOTPPPPNLPPOTPBPBPBPBPPPBPBPBPBPPPPPPPPPBPBPBPBNLPBPBOTPBOTPPDMDMDMPPPPPPPBPPPBPBOTDMPPPPPPPPPPPBPPDONLPPDOPBPBPBPBPBPBPBDOPPPPSHPPPBPBPBRMPBPBPBOTPBOTPPPPPBPPPPPBPPPPPPPPPPRMPPPPPPPBNLDONLDOOTPBPBPBPBPBPBDMDOPPNLPBPBPBPBDMPBOTPBPBPBOTPPPBDMPPDMRMPPPPPPPBPPPBPPOTPPPMRMPPPPPBPBDODOOTDMDOPPPBPBPPDONLNLDMDMDOPBPBPBPBPBDMDOPPPPPBPBDMPPPBDMPBPBPBPBPBPBPBPBDMPBPBOTPBDMPBDMPBPBPBPBPPPPPPPPPPPPPPDODOPPDMPPPPPBPPPBRMPBPBPBDOPPPPPBDMPPPBPBPBPBPBPBDMDMPBPBPPPBPBPPPPNLNLPPNLPBPBPBPBDMDMPPPBOTPBPBPPPBOTPBDMDOPBPBPPPBDOPPPBDMOTDMDMPPPPPPRMDOPPPPPBPBPBPBPPPBPPPBOTPPDMDMPBDODMPBPPPPOTPBDMNLPBPBPBPBPBPBPBPBPPNLPPDMDOPPPBPPPBOTPPDMDMDMPPOTPPPPDMPPPPPBDMPPPPDOPBPBDOPBPBPBPPPBPBPPOTOTPPPBPBPPPBDOPBPPPBPBNLDMOTPPOTPBPBPPPPPBOTPBPBDMDMPPPPDOPBPBPPNLPBPBPBPBPBPBPBPBPPPPPBPBPBPBPBPBPBPPPPPPPBPBDMPPPPPBPBPBPPPBPPPBPBPPPPOTPPPPPPDOPPPPPBOTPPPBDMPPPPPPPPPPOTPPPPPPPBDOPPPPPPPBPBPBPBPPPBOTPPPPPPPBPBPPPBPBPBPBPBPBPBPBPBPBPBPBPBPBPPPBPPNLPPPPDMOTDMPPDOPBPBPBPBPBPBPBPPPPPPPBPBPBOTPPPBPPDMPPOTPPPPPPPPPBOTPBPPPPDOPPPBPBPBPBPBOTPPDMPPPPPPPPPPPEPPPPPPPBPBPPPBPBPPDMPPPPPPPBPBPPPBDMPPPPDMPBPBPBPBPBPBPBPBPBPPPPPPPBPBPBPBPBPBPBPBPBPPPPPPPPPPPBPBPPPPPPPPOTDMDMPPPPPPPPNLPPPPPBPBPBPBPBPBPBPPPBPBPBPBPBPBPBPPPBPBPPDMOTOTDMPBPBPBPBPBPBPBPBPPPBPBPPPPPBPPPBPBOTDMDMDMPBPPOTPBPBPBPBPPDMPPDMOTPPOTPPPPDMOTPBPBPBPPDMOTPPPBOTDMNLPPPBPBOTDMDMPBDMOTOTPBPBNLPBPBPBPBPBPBPBPEPBOTPBDMPBPBPBPPOTPBPBDMPBPBDMPPPBOTPBPBPBDMDMOTDMOTPPPPOTOTOTPBPEDMNLOTDMOTNLPBDMDMDMPBPBPBPBPBNLPBOTDMOTOTPBPBDODMDMDMOTPBOTDOPBPBOTOTDMDMOTDMPBPBDMOTOTDMNLOTNLPBDMDMOTPBPBPBPBPBPBOTPBOTPBOTOTPBDMOTDMPBOTPBPBDMDMOTPBDMNLDMOTPBDMDMOTDMPBPBPBPBPBPBPBPBOTOTPMPBPBDMDMPBPBDMDMDMDMPBDMRMRMPBDMDMDMOTPBPBPBDMPBPBDMDMPPPBPBDMDMOTPBDMPPDMNLDMNLRMDMOTDMDMDMPBPBPBPBPBOTOTDMDMDMOTPMPPPBDMPPDMPPOTPPPBDOPPDMNLPPDOPPDMDOPPPEPBDMPPDMPBNLPBPBPBPFPBPBOTPBOTDMPPPBSHDMOTDMOTDMDMDMDOPPPFDMDOPMDMPFPPPPPPDONLDONLPBPBDMPBPBDMPPDMPBPBPBPBNLNLNLPBOTOTDMDMDMSHPBSHOTPBOTDMDMDMPPDMPBPPDOOTPPDMDMDMDMDMPPPPPFPBDOPPPPNLNLPPDMPPDONLDMPPPPNLPBPBPFPBPBPPPPDMDMDMDMPBPPPBPBPBPBPFPBPPNLPPPPPPPBPBPBPPDMPFPPPPPPPFPBPBPPPBPPPBPBPBPPPPPPPBDMPPDMDMPPDMDMDMDMPPPPPBPBPPPBDOPPPPPBPBOTPBPPDMDMDMDMOTPPPPPBPPPPNLPPPBPPDOPBDOPPDMPBOTOTNLPPDMPPPPPPPBPBPPPFPBPBPBPBPFPPDMPFDMPBPBPBPBOTPBPEPBPBPBPBPBPBPBPBPBPBPBPBPBOTPPPPPBPPPPPBPPPPPPPPPBDMDMPPPFPBPBPBPPPBPBPPPBPBPPPPPPPBPPPPDMOTPBDMDMDMPPPPOTPBPPPBPPPBDOPPPPPBOTPBPBOTPBDMDMDMDMPPDMPPPPPPPBPPPBPBPPPBDMPPPPDMPEPPPBPBPBPPOTPPPBPBPBPBPBPBPBPBDMDMPBPPDMPBPBNLPBPPPBPBPBPBPBPBPBPBOTPBPBPBPBOTPPPPPPPBPBPPDOPBOTPPDODMPFDMDMOTPBPBPPPBPBPBPBPBPBDMOTPPDMPBPBOTDMDMDMPPOTPPPBPBPPPBPBPBPPDMDMDMDMOTPPPPPBOTPBPBPBPBPBPBSHNLDMPBDMOTOTPPDMOTPBPBOTPPPBPBPFPBPBPBPBOTDMDMPBOTPBPBPBPBPBPBPBPBPBPBPBPEPBPBPBPEPBPEPBOTPBPPDOPBDMPBPFPFDMDOPBPFPBPBPBPBPBPBPPOTPBPBOTPBPBOTDMPPDMDMDMDMDMOTDMOTDOPBPPDOPBPBDOPBPBPFOTDMDMDMDMOTPPOTPBPBOTPBDODOPBPEDMPBPBNLNLDODODMOTPBPBPBPBPBPBPBPBPBDMDMPBNLPBPBPEPBNLNLPBPBPBPBPBPBPBPBPBOTPPRMOTRMPBPBOTPEPBPBPBDMPPDODMDMPBDODMDMPBPBPBPBOTOTDMDOPBPBDMDMDMDMPBOTPBPBPBDOPBOTOTPBDMDMOTDMOTPBOTDOPBPEDMPBNLNLDODMPBOTPBPEOTPBPBPBPBRMPBPBPBDMOTDMPBNLOTPBPBPBPBPBPBPBPBRMPEPBRMPBPBPBOTOTPEOTOTDODMPBPEDODODMPPPBPEOTPBPBPBPBPBOTPBDMDMPBPBPBPBPBPBDODODMSHPBPBOTPBDMDOPEPBPBDOPEDMDOPPDMPBDMNLRMPBPBPBPBPBPBPBOTPEPBDMPBPBPBPBPBPBPBPBOTPEDMDMDOPBPBPBPBPBPBPBDOPBNLDMDODMDMDMDMPBPBPBDODMDOPBPFOTPPDMOTDMDMPBRMRMRMPBDORMDORODODODODODMOTNLDOPBDOPBPBPBPBPBOTPBPBDMDMOTDMPBPBDMPBPBNLPBPBPBNLPBPBPBPBPBPBDMPBDOPFDMPBDOPBDMNLPBPBPBPBOTDOPBPBDMDMPPPBOTDODMDMPEPBDMPBPBDOPBDOOTDMDMOTDMPPPBRMDOPPDMPPPPPEPPOTPPPBPBDOPBPPDOPPPPPPDMDOPBDMDOPPDOPPPPDMPPPBDODOOTPBPBOTPBPBPPPBPBOTPBPBDMPBDODONLOTDMPPDMPPNLDMPBPBPBDOPPPBPPDODMPPPFPBOTPPDMDMDMPPOTDMPPOTDMDMDMPPPBPPPPPBPBPBPBPBPBPBPBPBNLPPPBDMDMNLDOPPPBPBDMPPNLOTPEPBPBDMPPDOPBPBPPNLDMPPPPPEDOPBPBPBPPPBPPPBPBPBPBPBPBDMPBPPDMPBPBPBOTNLPBPBPBPBPBPBPBPBPBPBPEPPPPPBDODMPBPBPBPFPPPPDMDMPPPPDMDOPBPBDODOPBPBPBSHPBPPPPPBPBPEPBDOPPDOPPDMPPOTDMDMDOPBDMDOPPPBPPPBPBPBDOOTPBPBDMDMPPPBPPPPPBDOPBPBPPPPDMDMPPPEPBNLDMDOPPPEPPPPOTPBPPPPPBPPPBPBPBPBPBPBPBPBPBPBPBPBPBPBDMPPPBPPDMPBDMPBPBPBPBPBPBPBPBPBPBPBPBNLRMPPPPPPDOPBDMPPPPPPPBPBPPPPPPDMDMPBPEPBPPPBPPPPPBPBDOPPDOPPDOPPOTPPDMDMPPPPDOPBPBPPDOPBDOPBPBPBDMDMPBPPPBPBPBPBPBPFOTPBPPDOPPDMPPPPPPPPPBPPPEPBDMPPPPPPDMPPPPNLPPPBPPPPPBPBOTPBPBPBPBPBPBPBPBPBPBPBPBPPPBDMPPPBPPDMPBPBPPPBPBPBPBPBPBPBPBPBPBPBPBPBPBDMPEOTPPPPPEPPNLPBPBOTPPOTPPPBPBPPPPDMPPPBDMPPPFPPPBPPPPPPPBPBPBPBPBPBPPPBPBPPDOPBPPPPDOOTPPPPPPOTPPPPDOPPPEDMPPPPDODMPPPPPPPBOTPPPPPBPPPBPPDMDMPPPBPPPBPBDMPPPBOTPPDMDOPPDMDODMNLPBPBPBPBDMPBPBNLPBPBPBDMOTPBDMDMOTPBDODOPPPPPPPPPPPBDOPBPPPBDOPPPBPPPBPPPBPBOTPBPPPPPPPBPPPPPPPPPPPPPBPPPPPBPBPBPPPBPBPPDMDMPPPBPBPBPBPBPBPBPBPBPBPBOTPEPBPBPPPBPPDOPPPBDOPPDMNLPBPBPBPBPBPPPBPBPBPBPBPBPBPBPBPBPBPBNLPEPBPBPBPBPBPBPBPFPPPBPBPBDOPPPBPBPBPBPBPBPPOTPPDODMPPPPPBOTPPDOPPDMPBPBPBPBPBPPPBPEPPDMDMDMDMPBPBPBPBDOPPPPPPPEPPOTPPPPPBPPPBOTPPPBOTPBPBPBPBPBPBPBPBPBPEPBPBPBPBPBPPOTPPPBPBPBDODMPBNLPBPBPBPBPBPBPBPBPBPBPBPBPBPBPBPEPBRMPBPEPBRMPEPBDOPBDMOTPBDODMOTPBPBPBPBPBPBPBDOPBPBPBPBPBPBPBPBPBOTPBPBPBPPPBPBPBPBDOOTDMDMDMPFPBPBDOPBPBPBPBDOPBPBOTOTDOOTPBDODONLPEPBPBPBPBDMPBPBPBPBPBPBPBPBPBPBPBNLPBPBPBPPDOPBDMPENLPBPBPBPBPBPBPBPBPBPBPBPBPBPPPBOTPEPBPBPBPBPBPBPBPBPBPBDOOTPFPBDOPBPBPBPBPBOTPBPBDMDOOTPBSFDOPBDMOTPBDOPBPBPBPBPBDOPBDMDMDMDMPBPBPEPBPBPBPBPBOTDODODOPPDMPBPBRONLPBPBDMDONLPBPBPBPBPBPBPBPBPBPBPBPBNLDMPBPBDMPBPBDMPBOTPBPBPBPBNLPBPBPBPBPBPBPBPBOTDOROPEPBPEPBPBOTDOPBPBPBOTPBPBPBSFDODMDMDMPBOTDOPBOTPBDMDMDMDMPBPBPBPBPBRM' to numeric
The above exception was the direct cause of the following exception:
TypeError Traceback (most recent call last)
Cell In[27], line 1
----> 1 grouped_data.mean()
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/groupby.py:2452, in GroupBy.mean(self, numeric_only, engine, engine_kwargs)
2445 return self._numba_agg_general(
2446 grouped_mean,
2447 executor.float_dtype_mapping,
2448 engine_kwargs,
2449 min_periods=0,
2450 )
2451 else:
-> 2452 result = self._cython_agg_general(
2453 "mean",
2454 alt=lambda x: Series(x, copy=False).mean(numeric_only=numeric_only),
2455 numeric_only=numeric_only,
2456 )
2457 return result.__finalize__(self.obj, method="groupby")
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/groupby.py:1998, in GroupBy._cython_agg_general(self, how, alt, numeric_only, min_count, **kwargs)
1995 result = self._agg_py_fallback(how, values, ndim=data.ndim, alt=alt)
1996 return result
-> 1998 new_mgr = data.grouped_reduce(array_func)
1999 res = self._wrap_agged_manager(new_mgr)
2000 if how in ["idxmin", "idxmax"]:
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/internals/managers.py:1469, in BlockManager.grouped_reduce(self, func)
1465 if blk.is_object:
1466 # split on object-dtype blocks bc some columns may raise
1467 # while others do not.
1468 for sb in blk._split():
-> 1469 applied = sb.apply(func)
1470 result_blocks = extend_blocks(applied, result_blocks)
1471 else:
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/internals/blocks.py:393, in Block.apply(self, func, **kwargs)
387 @final
388 def apply(self, func, **kwargs) -> list[Block]:
389 """
390 apply the function to my values; return a block if we are not
391 one
392 """
--> 393 result = func(self.values, **kwargs)
395 result = maybe_coerce_values(result)
396 return self._split_op_result(result)
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/groupby.py:1995, in GroupBy._cython_agg_general.<locals>.array_func(values)
1992 return result
1994 assert alt is not None
-> 1995 result = self._agg_py_fallback(how, values, ndim=data.ndim, alt=alt)
1996 return result
File ~/work/workshop-introduction-to-python/workshop-introduction-to-python/.venv/lib/python3.12/site-packages/pandas/core/groupby/groupby.py:1946, in GroupBy._agg_py_fallback(self, how, values, ndim, alt)
1944 msg = f"agg function failed [how->{how},dtype->{ser.dtype}]"
1945 # preserve the kind of exception that raised
-> 1946 raise type(err)(msg) from err
1948 if ser.dtype == object:
1949 res_values = res_values.astype(object, copy=False)
TypeError: agg function failed [how->mean,dtype->object]