import pandas as pd8 Combining DataFrames
Previously, we have seen how to analyze and manipulate data in a single DataFrame. However, you will often find data saved into different files and, therefore, you may need to deal with several different pandas DataFrames.
In this session we will explore different ways of combining DataFrames into a single DataFrame.
Let’s start loading the pandas library, reading two data sets into DataFrames, and having a quick look at the tabular data: surveys.csv and species.csv
surveys_df = pd.read_csv("../course_materials/data/surveys.csv", keep_default_na=False, na_values=[""])
species_df = pd.read_csv("../course_materials/data/species.csv", keep_default_na=False, na_values=[""])print(surveys_df.info())
surveys_df.head()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 35549 entries, 0 to 35548
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 record_id 35549 non-null int64
1 month 35549 non-null int64
2 day 35549 non-null int64
3 year 35549 non-null int64
4 plot_id 35549 non-null int64
5 species_id 34786 non-null object
6 sex 33038 non-null object
7 hindfoot_length 31438 non-null float64
8 weight 32283 non-null float64
dtypes: float64(2), int64(5), object(2)
memory usage: 2.4+ MB
None| record_id | month | day | year | plot_id | species_id | sex | hindfoot_length | weight | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 7 | 16 | 1977 | 2 | NL | M | 32.0 | NaN |
| 1 | 2 | 7 | 16 | 1977 | 3 | NL | M | 33.0 | NaN |
| 2 | 3 | 7 | 16 | 1977 | 2 | DM | F | 37.0 | NaN |
| 3 | 4 | 7 | 16 | 1977 | 7 | DM | M | 36.0 | NaN |
| 4 | 5 | 7 | 16 | 1977 | 3 | DM | M | 35.0 | NaN |
print(species_df.info())
species_df.head()<class 'pandas.core.frame.DataFrame'>
RangeIndex: 54 entries, 0 to 53
Data columns (total 4 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 species_id 54 non-null object
1 genus 54 non-null object
2 species 54 non-null object
3 taxa 54 non-null object
dtypes: object(4)
memory usage: 1.8+ KB
None| species_id | genus | species | taxa | |
|---|---|---|---|---|
| 0 | AB | Amphispiza | bilineata | Bird |
| 1 | AH | Ammospermophilus | harrisi | Rodent |
| 2 | AS | Ammodramus | savannarum | Bird |
| 3 | BA | Baiomys | taylori | Rodent |
| 4 | CB | Campylorhynchus | brunneicapillus | Bird |
We now have two DataFrames. The first, surveys_df, contains information on individuals of a species recorded in a survey, while the second, species_df, contains more detailed information on each species.
8.1 Concatenating DataFrames
The first way we will combine DataFrames is concatenation, i.e. simply putting DataFrames one after the other either vertically or horizontally.
Concatenation can be used if the DataFrames are similar, meaning that they either have the same rows or columns. We will see examples of this later.
To concatenate two DataFrames you will use the function pd.concat(), specifying as arguments the DataFrames to concatenate and axis=0 or axis=1 for vertical or horizontal concatenation, respectively.
We will only be looking at vertical concatenation, but it is good to be aware that there is more to be discovered beyond what we describe here.
Let us first obtain two small DataFrames from the larger surveys_df dataset.
# Subsetting DataFrames
surveys_df_sub_first10 = surveys_df.head(10)
surveys_df_sub_last10 = surveys_df.tail(10)We now have two DataFrames, one with the first ten rows of the original dataset, and another with the last ten rows.
Now, let us do some vertical concatenation or stacking. In this case the two DataFrames are simply stacked ‘on top of’ each other (remember to specifyaxis=0).
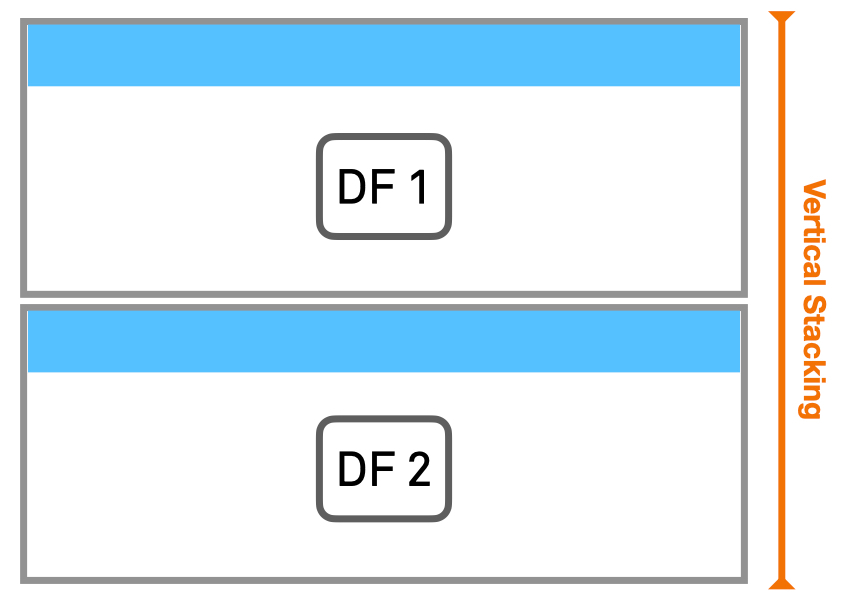
Vertical stacking can be understood as combining two DataFrames that have different sets of the same type of data. In our example, it may be that one field researcher has registered the first ten entries, and another did the last ten, both using the same laboratory sheets. They both wrote down the same information (weight, species, and so on) of all different individuals. If we combine them, we have one list of twenty records, rather than two lists of ten.
# Stack the DataFrames on top of each other
vertical_stack = pd.concat([surveys_df_sub_first10, surveys_df_sub_last10], axis=0)print(vertical_stack.info())
vertical_stack<class 'pandas.core.frame.DataFrame'>
Index: 20 entries, 0 to 35548
Data columns (total 9 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 record_id 20 non-null int64
1 month 20 non-null int64
2 day 20 non-null int64
3 year 20 non-null int64
4 plot_id 20 non-null int64
5 species_id 19 non-null object
6 sex 16 non-null object
7 hindfoot_length 15 non-null float64
8 weight 6 non-null float64
dtypes: float64(2), int64(5), object(2)
memory usage: 1.6+ KB
None| record_id | month | day | year | plot_id | species_id | sex | hindfoot_length | weight | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 7 | 16 | 1977 | 2 | NL | M | 32.0 | NaN |
| 1 | 2 | 7 | 16 | 1977 | 3 | NL | M | 33.0 | NaN |
| 2 | 3 | 7 | 16 | 1977 | 2 | DM | F | 37.0 | NaN |
| 3 | 4 | 7 | 16 | 1977 | 7 | DM | M | 36.0 | NaN |
| 4 | 5 | 7 | 16 | 1977 | 3 | DM | M | 35.0 | NaN |
| 5 | 6 | 7 | 16 | 1977 | 1 | PF | M | 14.0 | NaN |
| 6 | 7 | 7 | 16 | 1977 | 2 | PE | F | NaN | NaN |
| 7 | 8 | 7 | 16 | 1977 | 1 | DM | M | 37.0 | NaN |
| 8 | 9 | 7 | 16 | 1977 | 1 | DM | F | 34.0 | NaN |
| 9 | 10 | 7 | 16 | 1977 | 6 | PF | F | 20.0 | NaN |
| 35539 | 35540 | 12 | 31 | 2002 | 15 | PB | F | 26.0 | 23.0 |
| 35540 | 35541 | 12 | 31 | 2002 | 15 | PB | F | 24.0 | 31.0 |
| 35541 | 35542 | 12 | 31 | 2002 | 15 | PB | F | 26.0 | 29.0 |
| 35542 | 35543 | 12 | 31 | 2002 | 15 | PB | F | 27.0 | 34.0 |
| 35543 | 35544 | 12 | 31 | 2002 | 15 | US | NaN | NaN | NaN |
| 35544 | 35545 | 12 | 31 | 2002 | 15 | AH | NaN | NaN | NaN |
| 35545 | 35546 | 12 | 31 | 2002 | 15 | AH | NaN | NaN | NaN |
| 35546 | 35547 | 12 | 31 | 2002 | 10 | RM | F | 15.0 | 14.0 |
| 35547 | 35548 | 12 | 31 | 2002 | 7 | DO | M | 36.0 | 51.0 |
| 35548 | 35549 | 12 | 31 | 2002 | 5 | NaN | NaN | NaN | NaN |
The resulting DataFrame (vertical_stack) consists, as expected, of 20 rows. These are the result of the first and last 10 rows of our original DataFrame surveys_df.
You may have noticed that the last ten rows have very high index, not consecutive with the first ten rows. This is because concatenation preserves the indices of the two original DataFrames. If you want a brand new set of indices for your concateneted DataFrame, simply reset the indices using the method .reset_index(). Notice that this adds a column index to your DataFrame, that maintains the original index. If you pass drop=True into the function, you will avoid the addition of this column.
vertical_stack.reset_index()| index | record_id | month | day | year | plot_id | species_id | sex | hindfoot_length | weight | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 1 | 7 | 16 | 1977 | 2 | NL | M | 32.0 | NaN |
| 1 | 1 | 2 | 7 | 16 | 1977 | 3 | NL | M | 33.0 | NaN |
| 2 | 2 | 3 | 7 | 16 | 1977 | 2 | DM | F | 37.0 | NaN |
| 3 | 3 | 4 | 7 | 16 | 1977 | 7 | DM | M | 36.0 | NaN |
| 4 | 4 | 5 | 7 | 16 | 1977 | 3 | DM | M | 35.0 | NaN |
| 5 | 5 | 6 | 7 | 16 | 1977 | 1 | PF | M | 14.0 | NaN |
| 6 | 6 | 7 | 7 | 16 | 1977 | 2 | PE | F | NaN | NaN |
| 7 | 7 | 8 | 7 | 16 | 1977 | 1 | DM | M | 37.0 | NaN |
| 8 | 8 | 9 | 7 | 16 | 1977 | 1 | DM | F | 34.0 | NaN |
| 9 | 9 | 10 | 7 | 16 | 1977 | 6 | PF | F | 20.0 | NaN |
| 10 | 35539 | 35540 | 12 | 31 | 2002 | 15 | PB | F | 26.0 | 23.0 |
| 11 | 35540 | 35541 | 12 | 31 | 2002 | 15 | PB | F | 24.0 | 31.0 |
| 12 | 35541 | 35542 | 12 | 31 | 2002 | 15 | PB | F | 26.0 | 29.0 |
| 13 | 35542 | 35543 | 12 | 31 | 2002 | 15 | PB | F | 27.0 | 34.0 |
| 14 | 35543 | 35544 | 12 | 31 | 2002 | 15 | US | NaN | NaN | NaN |
| 15 | 35544 | 35545 | 12 | 31 | 2002 | 15 | AH | NaN | NaN | NaN |
| 16 | 35545 | 35546 | 12 | 31 | 2002 | 15 | AH | NaN | NaN | NaN |
| 17 | 35546 | 35547 | 12 | 31 | 2002 | 10 | RM | F | 15.0 | 14.0 |
| 18 | 35547 | 35548 | 12 | 31 | 2002 | 7 | DO | M | 36.0 | 51.0 |
| 19 | 35548 | 35549 | 12 | 31 | 2002 | 5 | NaN | NaN | NaN | NaN |
8.2 Joining DataFrames
Concatenating DataFrames allows you to combine two entire DataFrames into a single one. In many cases, you want to combine only selected parts of two DataFrames.
You might, for example, want to merge rows of two DataFrames that have matching values in specific columns. The pandas function merge() performs an operation that you may know as a join if you worked with databases before. The join operation joins the content of two DataFrames in a particular way. There are different types of joins, but the workflow to perform a join operation is always the same:
- You identify a left and a right DataFrame, among the two you want to join;
- You identify in both your left and right DataFrame a column (or set of columns) to join on;
- You choose the type of join;
- You perform the join running the function
pd.merge()with the specified inputs and options.
What it means for a DataFrame to be ‘left’ or ‘right’ depends on the type of join. The main thing to remember is that it matters which DataFrame you mention first when performing a join.
We will only be looking at what is know as an inner join. Investigating the other types of joins is left as an exercise.
Let’s see some join example using the first ten rows of our surveys_df DataFrame (surveys_df_sub_first10). This will be our left DataFrame, containing data on individual animals. Our right DataFrame will be the first 20 rows of species_df, containing some information on each species. Let us rename them for the purpose of the join:
left_df = surveys_df_sub_first10
right_df = species_df.head(20)left_df| record_id | month | day | year | plot_id | species_id | sex | hindfoot_length | weight | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 7 | 16 | 1977 | 2 | NL | M | 32.0 | NaN |
| 1 | 2 | 7 | 16 | 1977 | 3 | NL | M | 33.0 | NaN |
| 2 | 3 | 7 | 16 | 1977 | 2 | DM | F | 37.0 | NaN |
| 3 | 4 | 7 | 16 | 1977 | 7 | DM | M | 36.0 | NaN |
| 4 | 5 | 7 | 16 | 1977 | 3 | DM | M | 35.0 | NaN |
| 5 | 6 | 7 | 16 | 1977 | 1 | PF | M | 14.0 | NaN |
| 6 | 7 | 7 | 16 | 1977 | 2 | PE | F | NaN | NaN |
| 7 | 8 | 7 | 16 | 1977 | 1 | DM | M | 37.0 | NaN |
| 8 | 9 | 7 | 16 | 1977 | 1 | DM | F | 34.0 | NaN |
| 9 | 10 | 7 | 16 | 1977 | 6 | PF | F | 20.0 | NaN |
right_df| species_id | genus | species | taxa | |
|---|---|---|---|---|
| 0 | AB | Amphispiza | bilineata | Bird |
| 1 | AH | Ammospermophilus | harrisi | Rodent |
| 2 | AS | Ammodramus | savannarum | Bird |
| 3 | BA | Baiomys | taylori | Rodent |
| 4 | CB | Campylorhynchus | brunneicapillus | Bird |
| 5 | CM | Calamospiza | melanocorys | Bird |
| 6 | CQ | Callipepla | squamata | Bird |
| 7 | CS | Crotalus | scutalatus | Reptile |
| 8 | CT | Cnemidophorus | tigris | Reptile |
| 9 | CU | Cnemidophorus | uniparens | Reptile |
| 10 | CV | Crotalus | viridis | Reptile |
| 11 | DM | Dipodomys | merriami | Rodent |
| 12 | DO | Dipodomys | ordii | Rodent |
| 13 | DS | Dipodomys | spectabilis | Rodent |
| 14 | DX | Dipodomys | sp. | Rodent |
| 15 | EO | Eumeces | obsoletus | Reptile |
| 16 | GS | Gambelia | silus | Reptile |
| 17 | NL | Neotoma | albigula | Rodent |
| 18 | NX | Neotoma | sp. | Rodent |
| 19 | OL | Onychomys | leucogaster | Rodent |
The column we want to perform the join on is the one containing information about the species_id. Conveniently, this column has the same label in both DataFrames. Note that this is not always the case.
inner_join = pd.merge(left_df, right_df, left_on='species_id', right_on='species_id', how='inner')
inner_join| record_id | month | day | year | plot_id | species_id | sex | hindfoot_length | weight | genus | species | taxa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 7 | 16 | 1977 | 2 | NL | M | 32.0 | NaN | Neotoma | albigula | Rodent |
| 1 | 2 | 7 | 16 | 1977 | 3 | NL | M | 33.0 | NaN | Neotoma | albigula | Rodent |
| 2 | 3 | 7 | 16 | 1977 | 2 | DM | F | 37.0 | NaN | Dipodomys | merriami | Rodent |
| 3 | 4 | 7 | 16 | 1977 | 7 | DM | M | 36.0 | NaN | Dipodomys | merriami | Rodent |
| 4 | 5 | 7 | 16 | 1977 | 3 | DM | M | 35.0 | NaN | Dipodomys | merriami | Rodent |
| 5 | 8 | 7 | 16 | 1977 | 1 | DM | M | 37.0 | NaN | Dipodomys | merriami | Rodent |
| 6 | 9 | 7 | 16 | 1977 | 1 | DM | F | 34.0 | NaN | Dipodomys | merriami | Rodent |
As you may notice, the resulting DataFrame has only seven rows, while our original DataFrames had 10 and 20 rows, respectively. This is because an inner join selects only those rows where the value of the joined column occurs in both DataFrames (mathematically, an intersection).
Aside from the inner join, there are three more types of join that you can do using the merge() function: - An inner join selects only the rows that result from the combination of matching rows in both the original left and right DataFrames (intersection); - A left join selects all rows that were in the original left DataFrame, some of which may have been joined with a matching entry from the right DataFrame; - A right join selects all rows that were in the original right DataFrame, some of which may have been joined with a matching entry from the left DataFrame; - An outer join merges the two DataFrames and keeps all resulting rows.
To better understand how join works, it may be useful to look at the diagrams below:
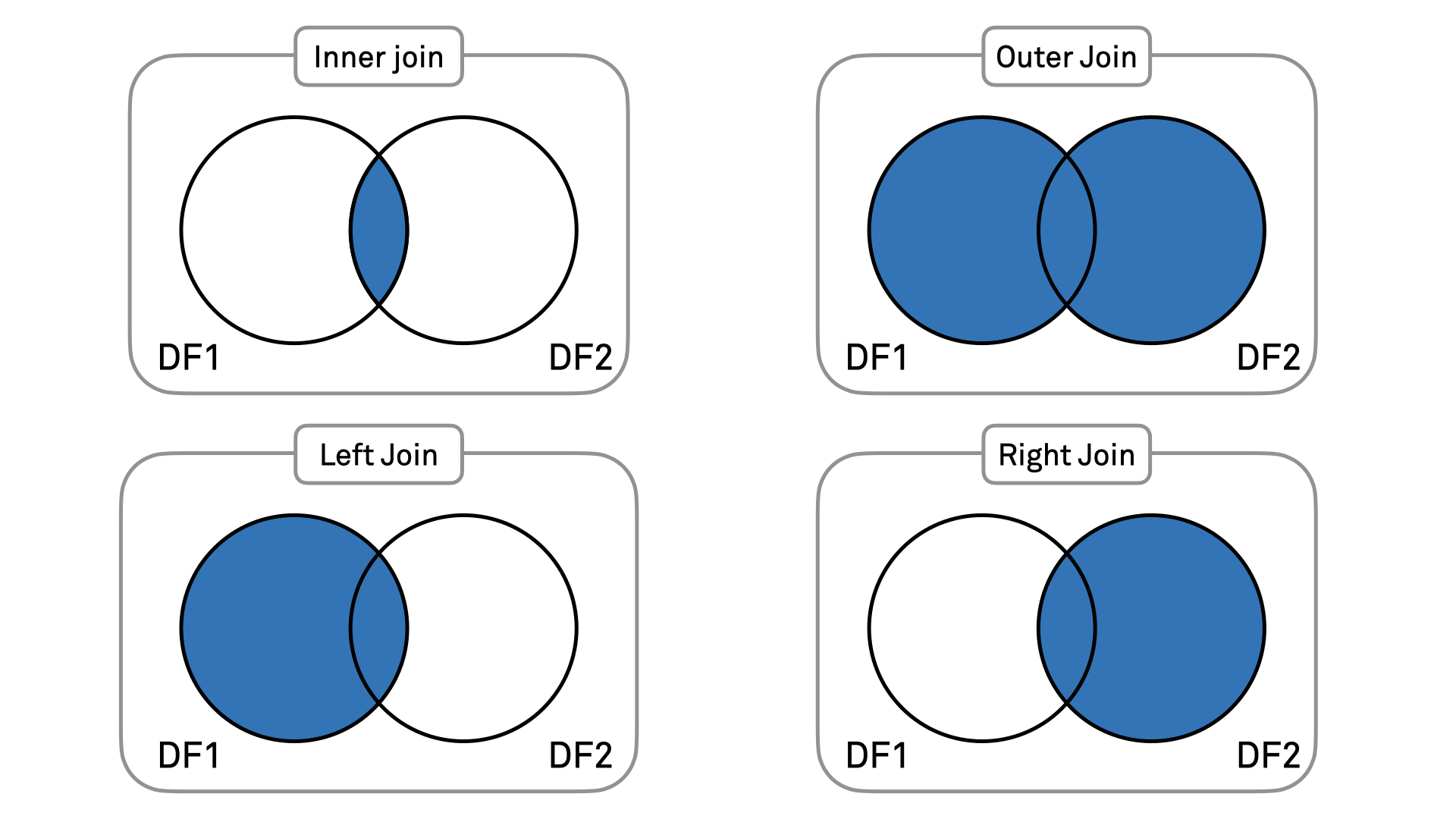
- Do you want to select only common information between the two DataFrames? Then you use an inner join;
- Do you want to add information to your left DataFrame? Then you use a left join;
- Do you want to add information to your right DataFrame? Then you use a right join;
- Do you want to get all the information from the two DataFrames? Then you use an outer join.
Exercises 9 and 10
Now go to the Jupyter Dashboard in your internet browser and continue with exercise 9 and 10.
We will continue with Data visualization.
